The molecular basis of lamin-specific chromatin interactions
- PMID: 40750945
- PMCID: PMC12527912
- DOI: 10.1038/s41594-025-01622-5
The molecular basis of lamin-specific chromatin interactions
Abstract
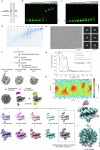
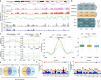
In the cell nucleus, chromatin is anchored to the nuclear lamina, a network of lamin filaments and binding proteins that underly the inner nuclear membrane. The nuclear lamina is involved in chromatin organization through the interaction of lamina-associated domains within the densely packed heterochromatin regions. Using cryo-focused ion beam milling in conjunction with cryo-electron tomography, we analyzed the distribution of nucleosomes at the lamin-chromatin interface at the nanometer scale. Depletion of lamins A and C reduced nucleosome concentration at the nuclear periphery, while B-type lamin depletion contributed to nucleosome density in proximity to the lamina but not further away. We then investigated whether specific lamins can mediate direct interactions with chromatin. Using cryo-electron microscopy, we identified a specific binding motif of the lamin A tail domain that interacts with nucleosomes, distinguishing it from the other lamin isoforms. Furthermore, we examined chromatin structure dynamics using a genome-wide analysis that revealed lamin-dependent macroscopic-scale alterations in gene expression and chromatin remodeling. Our findings provide detailed insights into the dynamic and structural interplay between lamin isoforms and chromatin, molecular interactions that shape chromatin architecture and epigenetic regulation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures














References
-
- Burke, B. & Stewart, C. L. The nuclear lamins: flexibility in function. Nat. Rev. Mol. Cell Biol.14, 13–24 (2013). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

