Artificial intelligence for prediction of atrial fibrillation in the stroke unit: a retrospective derivation validation cohort study
- PMID: 40752407
- PMCID: PMC12341230
- DOI: 10.1016/j.ebiom.2025.105869
Artificial intelligence for prediction of atrial fibrillation in the stroke unit: a retrospective derivation validation cohort study
Abstract
Background: Paroxysmal atrial fibrillation (AF) is a major cause of stroke but is often undetected in routine clinical practice. Effective stratification is needed to identify patients with stroke who might benefit the most from intensified AF screening. Several artificial intelligence models have been proposed to predict AF based on ECG in sinus rhythm, but broad implementation has been limited. The most valuable input features and most effective model design for AF prediction are also unclear.
Methods: We developed and tested AF prediction models utilising continuous electrocardiogram monitoring (CEM) recordings from the first 72 h after admission and multiple clinical input features from patients with stroke hospitalised at Charité, Berlin, Germany, between September 2020 and August 2023. We compared different models and input data to identify the best-performing model for prediction of AF. The relative contributions of different input data sources were assessed for explainability. A final model was externally validated using the first hour of monitoring data from the intervention group of the prospective multicentre MonDAFIS study.
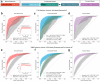
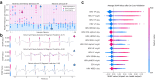
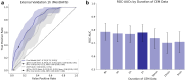
Findings: The derivation dataset included 2068 patients with acute ischaemic stroke, of whom 469 (22.7%) had AF, first detected before or during the index hospital stay (366 vs. 103). In predicting newly detected AF, a Bayesian fusion model emerged as best, achieving a ROC-AUC of 0.89 (95% CI: 0.80, 0.96). Model introspection indicated that HRV was the main driver of the model's predictions. A final, simplified tree-based ensemble model using age and HRV parameters of the first hour of CEM data achieved similar performance (ROC-AUC 0.88, 95% CI: 0.79, 0.95). The final model consistently outperformed the AS5F score in a real-world scenario external validation on the MonDAFIS dataset (1519 patients, thereof 36 (2.37%) with AF; ROC-AUC 0.79 vs. ROC-AUC 0.69, p = 4.69e-03).
Interpretation: HRV appears to be the most informative variable for predicting AF. A computationally inexpensive model requiring only 1 h of single-lead CEM data and patients' age supports prediction of AF after acute ischaemic stroke for up to seven days. Such a model may enable risk-based stratification for cardiac monitoring, prioritising efforts where most needed to enhance AF screening efficiency and, ultimately, secondary stroke prevention.
Funding: This study was supported by the German Federal Ministry of Education and Research and the German Research Foundation.
Keywords: Artificial intelligence; Atrial fibrillation; Heart rate variability; Machine learning; Prediction; Stroke.
Copyright © 2025 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests CHN received speaker and/or consultation fees from Alexion, Astra Zeneca, Bristol-Myers Squibb, Novartis, Pfizer Pharma, and Bayer, all outside the submitted work. JFS received research grants from the German Heart Foundation and speaker/consultation honoraria from Medtronic, Astra Zeneca, Bayer, and Bristol-Myers Squibb, outside the submitted work. MGK received a grant from the German Heart Foundation. AM received research grants from the German Research Foundation and the Leducq Foundation. KGH reports speaker's honoraria, consulting fees, lecture honoraria and/or study grants from Bayer, AstraZeneca, Pfizer, Bristol-Meyer-Squibb, Daiichi Sankyo, Boehringer Ingelheim, and Novartis. ME reports grants from Bayer and Ipsen and fees paid to the Charité from Amgen, AstraZeneca, Bayer, BMS, Daiichi Sankyo, all outside the submitted work. The remaining authors have no conflicts of interest or competing interests to declare.
Figures


References
-
- Kolmos M., Christoffersen L., Kruuse C. Recurrent ischemic stroke – a systematic review and meta-analysis. J Stroke Cerebrovasc Dis. 2021;30(8) - PubMed
-
- Sposato L.A., Lip G.Y.H., Haeusler K.G. Atrial fibrillation first detected after stroke: is timing and detection intensity relevant for stroke risk? Eur Heart J. 2024;45(5):396–398. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

