A mucosal vaccine prevents eosinophilic allergic airway inflammation by modulating immune responses to allergens in a murine model of airway disease
- PMID: 40753200
- PMCID: PMC12317988
- DOI: 10.1038/s41467-025-62632-x
A mucosal vaccine prevents eosinophilic allergic airway inflammation by modulating immune responses to allergens in a murine model of airway disease
Abstract
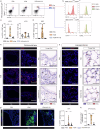
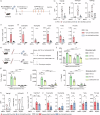
Allergic sensitization and viral infections are risk factors for asthma development and progression. Sublingual vaccination with MV130, a whole heat-inactivated polybacterial preparation, protects against viral infections, but its impact on allergic sensitization and asthma development remains unknown. Here we show MV130 prevents house dust mite (HDM)-induced local type 2 immune responses and associated eosinophilic airway inflammation, conferring protection up to 9 weeks after vaccination. MV130 reduces pathophysiological and clinical asthma features in an in vivo experimental mouse model of HDM-induced allergic eosinophilic asthma, restoring normal airway functionality. MV130 impairs allergen-specific IgE sensitization and systemic type 2 inflammation endorsing type 1 and IL-10 responses. In human DCs, MV130 induces a transcriptomic and metabolic reprogramming, and restores non-pathological immune responses to allergens in healthy and asthmatic donors. Additionally, the adoptive transfer of MV130-stimulated BMDCs was sufficient to reproduce the protective features of the vaccine administration in vivo. Collectively, we show MV130 reduces allergic sensitization and eosinophilic asthma. Our findings support the exploration of mucosal interventions aimed at reducing the risk of allergen-induced asthma development.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: O.P. received research grants from MINECO, Ministerio de Ciencia e Innovación, Inmunotek S.L., Novartis, and AstraZeneca and fees for giving scientific lectures or participation in Advisory Boards from: AstraZeneca, Pfizer, GlaxoSmithKline, Inmunotek S.L, Novartis, Sanofi-Genzyme and Regeneron. J.L.S. is the founder and CEO of Inmunotek SL and C.S.-O. and L.C. are employees of Inmunotek S.L. The rest of authors have no conflict of interest to declare.
Figures






References
-
- Venkatesan, P. 2023 GINA report for asthma. Lancet Respir. Med.11, 589 (2023). - PubMed
-
- Agache, I. et al. EAACI biologicals guidelines-recommendations for severe asthma. Allergy76, 14–44 (2021). - PubMed
-
- Kolkhir, P. et al. Type 2 chronic inflammatory diseases: targets, therapies and unmet needs. Nat. Rev. Drug Discov.22, 743–767 (2023). - PubMed
-
- Akdis, C. A. et al. Type 2 immunity in the skin and lungs. Allergy75, 1582–1605 (2020). - PubMed
-
- Wenzel, S. E. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat. Med.18, 716–25 (2012). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

