Integrating genomic and pathological characteristics to enhance prognostic precision in advanced NSCLC
- PMID: 40753345
- PMCID: PMC12318087
- DOI: 10.1038/s41698-025-01056-8
Integrating genomic and pathological characteristics to enhance prognostic precision in advanced NSCLC
Abstract
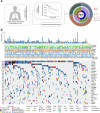
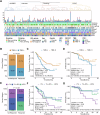
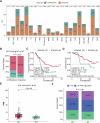
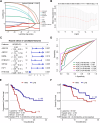
Although immunotherapy combined with chemotherapy (ICT) is the standard treatment for advanced non-small cell lung cancer (NSCLC), identification of reliable prognostic biomarkers remains challenging. In this multicenter study, we performed next-generation sequencing of tumor samples from 162 patients receiving first-line ICT at the Chinese PLA General Hospital and collected their pathological image information. First, we established a model to predict the risk of tumor progression based on genomic characteristics. Furthermore, a deep learning method was employed to recognize different cell types from pathological images, which significantly improved the accuracy of progression-free survival (PFS) and overall survival (OS) prediction. In summary, we constructed a Prognostic Multimodal Classifier for Progression (PMCP) that possesses the capability to precisely forecast PFS and OS. Patients with the PMCP1 subtype exhibit a low risk of progression and demonstrate a higher proportion of epithelial cells. PMCP highlighted the potential value of multimodal biomarkers in guiding clinical decisions regarding ICT. The area under curve (AUC) for predicting PFS was 0.807. This study revealed the importance of integrating genomic and pathological data to improve prognostic accuracy and enable personalized treatment for patients with advanced NSCLC.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare the following competing interests: S.Y.L., J.L.Z., Y.Z., J.Y.S. and J.C. are employed by Beijing ChosenMed Clinical Laboratory Co., Ltd. but declare no non-financial competing interests. All other authors declare no financial or non-financial competing interests.
Figures


References
-
- Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.74, 229–263 (2024). - PubMed
-
- Siegel, R. L., Miller, K. D., Wagle, N. S. & Jemal, A. Cancer statistics, 2023. Cancer J. Clin.73, 17–48 (2023). - PubMed
-
- SEER*Explorer: An interactive website for SEER cancer statistics [Internet]. Surveillance Research Program, National Cancer Institute; 2025 Jul 2. [cited 2025 Jul 22]. Available from: https://seer.cancer.gov/statistics-network/explorer/.
-
- Davodabadi, F. et al. Cancer chemotherapy resistance: mechanisms and recent breakthrough in targeted drug delivery. Eur. J. Pharmacol.958, 176013 (2023). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

