Multi-omics integration analysis identifies INPP4B as a T-cell-specific activation suppressor
- PMID: 40754682
- PMCID: PMC12318829
- DOI: 10.1002/ctm2.70430
Multi-omics integration analysis identifies INPP4B as a T-cell-specific activation suppressor
Abstract
Background: Naïve T cells are maintained in a quiescent state prior to activation. As inappropriate T-cell activation can lead to impaired immune tolerance and autoimmune diseases, the transition from quiescence to activation must be under strict regulation. Despite its importance, the mechanisms underlying the maintenance of the quiescent state remain incompletely understood.
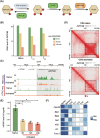
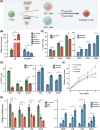
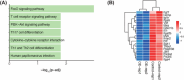
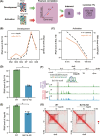
Methods and results: Through multi-omics integration analysis, we reveal that INPP4B, a phosphatase of the phosphoinositide 3-kinase pathway, is highly expressed specifically in T cells and is involved in suppressing T-cell activation and maintaining quiescence. Our findings uncover that INPP4B forms a T-cell-specific chromatin interaction domain and exhibits high expression levels in quiescent T cells. Upon T-cell activation, both the chromatin interaction and expression levels of INPP4B decrease. Functional studies further confirm that INPP4B suppresses T-cell activation and effector functions. Additionally, we observe increased expression level of INPP4B in exhausted T cells within the tumour microenvironment.
Conclusion: These results highlight the importance of maintaining optimal levels of INPP4B for T-cell function. Our findings suggest that INPP4B could be a potential target for enhancing the efficacy of T-cell-mediated immune responses against tumours.
Key points: A comprehensive multi-omics analysis characterizes the expression patterns of INPP4B across immune populations. INPP4B exhibits a T-cell-specific expression domain and functions as a T cell activation suppressor. INPP4B is significantly upregulated in exhausted T cells within the tumour microenvironment.
Keywords: INPP4B; T‐cell activation; multi‐omics.
© 2025 The Author(s). Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare they have no conflicts of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
