The Role of HINT3 in Myocardial Ischemia-Reperfusion Injury in Male Mice: Mechanisms Involving SDHA and its Acetylation
- PMID: 40755357
- PMCID: PMC12412524
- DOI: 10.1002/advs.202503109
The Role of HINT3 in Myocardial Ischemia-Reperfusion Injury in Male Mice: Mechanisms Involving SDHA and its Acetylation
Abstract
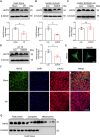
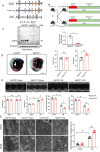
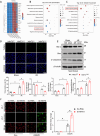
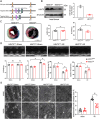
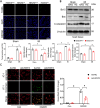
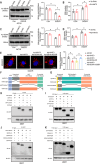
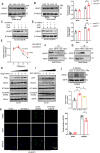
Myocardial ischemia-reperfusion (I/R) injury is characterized by oxidative stress, mitochondrial dysfunction, and cardiomyocyte apoptosis. During I/R, the accumulation and oxidation of succinate contribute to reactive oxygen species (ROS) production, worsening tissue damage. Histidine triad nucleotide-binding protein 3 (HINT3) is identified as a regulator of mitochondrial function and cardiomyocyte survival during I/R. In a mouse I/R model and an oxygen-glucose deprivation/reoxygenation (OGD/R) model, it shows that HINT3 expression is downregulated after I/R. Cardiomyocyte-specific knockout of HINT3 exacerbates myocardial injury, impairs cardiac function, and promotes mitochondrial dysfunction and apoptosis, whereas HINT3 overexpression mitigates these effects. Mechanistically, HINT3 interacts with succinate dehydrogenase subunit A (SDHA), suppresses HDAC1 expression, and prevents SDHA deacetylation at K335, reducing SDH activity and mitochondrial ROS production. These findings highlight the HINT3-HDAC1-SDHA axis as a key pathway in mitochondrial regulation, offering new insights and therapeutic targets for myocardial reperfusion injury.
Keywords: HINT3; SDHA; acetylation; mitochondrial dysfunction; myocardial ischemia/reperfusion.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Welt F. G. P., Batchelor W., Spears J. R., Penna C., Pagliaro P., Ibanez B., Drakos S. G., Dangas G., Kapur N. K., Journal of the American College of Cardiology 2024, 83, 2196. - PubMed
-
- Hansen P. R., Circulation 1995, 91, 1872. - PubMed
-
- Gonnot F., Boulogne L., Brun C., Dia M., Gouriou Y., Bidaux G., Chouabe C., Da Silva C. C., Ducreux S., Pillot B., Kaczmarczyk A., Leon C., Chanon S., Perret C., Sciandra F., Dargar T., Gache V., Farhat F., Sebbag L., Bochaton T., Thibault H., Ovize M., Paillard M., Gomez L., Nat. Commun. 2023, 14, 3346. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
