Targeting the BAG2/CHIP axis promotes gastric cancer apoptosis by blocking apoptosome assembly
- PMID: 40755756
- PMCID: PMC12313710
- DOI: 10.3389/fimmu.2025.1578416
Targeting the BAG2/CHIP axis promotes gastric cancer apoptosis by blocking apoptosome assembly
Abstract
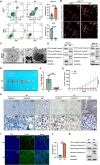
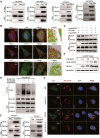
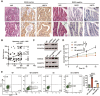
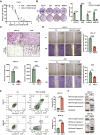
Apoptosis has been shown to play an important role in the treatment of gastric cancer, and BCL2-associated athanogene 2(BAG2) has been found to be able to inhibit apoptosis by interacting with multiple apoptosis regulators. In this study, we demonstrate that BAG2 functions as an independent prognostic factor, correlating with unfavorable clinical outcomes in patients with gastric cancer (GC). We demonstrate that BAG2 upregulation inhibited apoptosis and increased proliferation, migration, and invasion of GC cells, whereas the opposite results were obtained in BAG2-deficient GC cells. Mechanistically, BAG2 interacts with the c-terminus of HSP70-interacting protein(CHIP) to inhibit the ubiquitination degradation of Heat shock protein70(HSP70) and increase the binding of HSP70 to apoptotic protease-activating factor 1(Apaf1). The reduced ubiquitination degradation of HSP70 reduces the release of mitochondrial cytochrome C (Cytc), which ultimately inhibits the formation of apoptotic bodies assembled by Cytc and Apaf1. The above effects of BAG2 inhibit the formation of Cytc and Apaf1-assembled apoptotic bodies. Furthermore, we screened FIIN-2, an inhibitor of the BAG2 complex, which effectively halts the malignant development of GC triggered by reduced apoptosis by blocking BAG-CHIP binding. In conclusion, this study highlights BAG2's key role in regulating apoptosis and confirms FIIN-2's effectiveness in GC-targeted therapy.
Keywords: BAG2; FIIN-2; apoptosis; apoptosome; gastric cancer.
Copyright © 2025 Liu, Chen, Wei, Tang, Tian, Ma, Gu, Su, Dong, Shi and Luo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
BAG2 Inhibits Cervical Cancer Progression by Modulating Type I Interferon Signaling through Stabilizing STING.Adv Sci (Weinh). 2025 Aug;12(29):e70005. doi: 10.1002/advs.202414637. Epub 2025 May 14. Adv Sci (Weinh). 2025. PMID: 40364789
-
MYBL2 promotes cell proliferation and inhibits cell apoptosis via PI3K/AKT and BCL2/BAX/Cleaved-caspase-3 signaling pathway in gastric cancer cells.Sci Rep. 2025 Mar 17;15(1):9148. doi: 10.1038/s41598-025-93022-4. Sci Rep. 2025. PMID: 40097530 Free PMC article.
-
UBE4B promotes gastric cancer proliferation and metastasis by mediating FAT4 ubiquitination and degradation.Cell Death Dis. 2025 Jul 23;16(1):551. doi: 10.1038/s41419-025-07794-8. Cell Death Dis. 2025. PMID: 40701960 Free PMC article.
-
Chemotherapy for advanced gastric cancer.Cochrane Database Syst Rev. 2017 Aug 29;8(8):CD004064. doi: 10.1002/14651858.CD004064.pub4. Cochrane Database Syst Rev. 2017. PMID: 28850174 Free PMC article.
-
Molecular-targeted first-line therapy for advanced gastric cancer.Cochrane Database Syst Rev. 2016 Jul 19;7(7):CD011461. doi: 10.1002/14651858.CD011461.pub2. Cochrane Database Syst Rev. 2016. PMID: 27432490 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

