Molecular Characterization and Genetic Diversity of Broad Bean Wilt Virus 2 (BBWV-2) from
- PMID: 40757346
- PMCID: PMC12311748
- DOI: 10.1021/acsomega.5c04600
Molecular Characterization and Genetic Diversity of Broad Bean Wilt Virus 2 (BBWV-2) from
Abstract
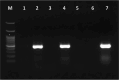
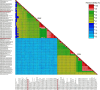
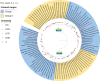
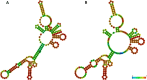
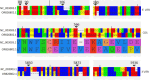
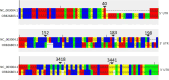
Small RNA deep sequencing (sRNA-seq) is a powerful tool for detecting known and novel plant viruses by analyzing virus-derived small RNAs (vsRNAs). In this study, the complete genome of Broad Bean Wilt Virus 2 (BBWV-2) was recovered from leaf samples using sRNA-seq, followed by bioinformatics analysis. The Genome Detective Tool, an online bioinformatics platform, identified both RNA1 (5.952 kb) and RNA2 (3.509 kb) of the BBWV-2 form raw data. RNA1 and RNA2 showed 78.8% and 78.1% nucleotide identity, and 88.2% and 87.4% amino acid identity, respectively, with their corresponding reference sequences. Comparative sequence analysis identified multiple mutations, comprising 8 and 6 indels distributed across the full-length sequences of RNA1 and RNA2, respectively, along with 1027 and 690 single-nucleotide substitutions localized within their coding regions. Analysis of the 5' untranslated regions (UTRs) identified five distinct motifs from both RNAs. Structural modeling suggested that RNA2 exhibits greater flexibility than RNA1, which may influence its biological functions. Phylogenetic analysis placed the Indian isolate within Group I, clustering with isolates from South Korea, China, the USA, and Germany, without clear host or geographic clustering. This study provides the first complete sequences of BBWV-2 RNA1 and RNA2 from India and represents the first global report of its occurrence in . Further, these findings enhance our understanding of the molecular diversity and evolutionary dynamics of BBWV-2.
© 2025 The Authors. Published by American Chemical Society.
Figures


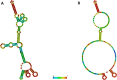
Similar articles
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Identification of a Broad Bean Wilt Virus 2 (BBWV2) Isolate (BBWV2-SP) from Spinacia oleracea L.Int J Mol Sci. 2025 Jun 20;26(13):5946. doi: 10.3390/ijms26135946. Int J Mol Sci. 2025. PMID: 40649722 Free PMC article.
-
Occurrence, distribution, and genetic diversity of faba bean viruses in China.Front Microbiol. 2024 Jun 19;15:1424699. doi: 10.3389/fmicb.2024.1424699. eCollection 2024. Front Microbiol. 2024. PMID: 38962134 Free PMC article.
-
Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article.
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
References
-
- Rui P. H., Jiang L., Li S., Jiang X. Z., Zhao Q. Q., Feng J. Y., Jiang T.. First Report of Broad Bean Wilt Virus 2 Infection in Eggplant in China. J. Plant Pathol. 2020;102:543. doi: 10.1007/s42161-019-00434-z. - DOI
-
- Gang S., Zhang S., Zhang S., Sun J., Yang C.. Broad Bean Wilt Virus 2 in Commelina communis L. in China. Bangladesh J. Bot. 2023;52:569–574. doi: 10.3329/bjb.v52i20.68222. - DOI
LinkOut - more resources
Full Text Sources
