Trichothiodystrophy-causative pathogenic variants impair a cooperative action of TFIIH and DDX1 in R-loop processing
- PMID: 40757642
- PMCID: PMC12319539
- DOI: 10.1093/nar/gkaf745
Trichothiodystrophy-causative pathogenic variants impair a cooperative action of TFIIH and DDX1 in R-loop processing
Abstract
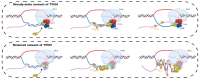
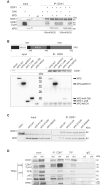
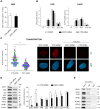
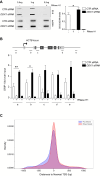
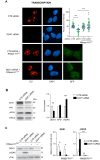
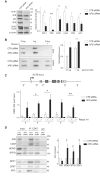
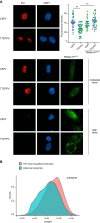
The transcription factor IIH (TFIIH) is a key player in transcription and DNA repair by nucleotide excision repair. It is made of 10 subunits organized in core-TFIIH and CAK sub-complexes bridged by XPD. Pathogenic variants in the ERCC2/XPD gene give rise to xeroderma pigmentosum (XP) or trichothiodystrophy (TTD), two distinct clinical entities with opposite skin cancer proneness. Here, we show that TTD variants cause a partial dissociation of the CAK from the chromatin and from the core-TFIIH. Mass spectrometry analysis reveals that the chromatin-bound CAK, as a component of the entire TFIIH, participates in a protein assembly containing the RNA-binding proteins DDX1, SFPQ, NONO as well as RNA polymerase II (Pol II). Gene silencing experiments demonstrate that the protein assembly is required to process the DNA:RNA hybrids formed during Pol II extension and to protect the cell from transcriptional stress. TTD-specific variants in ERCC2/XPD result in TFIIH instability, altered interaction of the CAK with DDX1-SFPQ-NONO, and R-loop accumulation. Therefore, the limited amount of TFIIH that distinguishes TTD from XP gives rise to transcriptional stress and extensive gene expression deregulations, thus accounting for the wide spectrum of TTD clinical features.
© The Author(s) 2025. Published by Oxford University Press.
Conflict of interest statement
None declared.
Figures


References
-
- Kolesnikova O, Radu L, Poterszman A TFIIH: a multi-subunit complex at the cross-roads of transcription and DNA repair. Adv Protein Chem Struct Biol. 2019; 115:21–67. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

