Differential gene reactions reveal drought response strategies in African acacias
- PMID: 40758440
- PMCID: PMC12320966
- DOI: 10.1111/tpj.70385
Differential gene reactions reveal drought response strategies in African acacias
Abstract
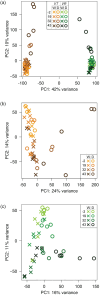
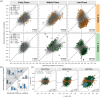
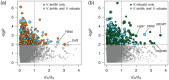
Drought tolerance involves a complex series of genetic reactions that expand over time as water stress intensifies. We investigated gene expression reactions over 43 days of drought stress in two widespread African savanna trees: the umbrella acacia (Vachellia tortilis) and the splendid thorn acacia (Vachellia robusta). Using 80 transcriptomes from droughted and watered individuals over time, we developed and implemented an analytical approach to identify genes with different reactions between a watered control and droughted treatment population of each species while filtering out genes changing similarly in both populations as part of normal growth and development. Our results show that both species use similar genetic systems to modulate photosynthesis, redox homeostasis, and hormone signaling, but they activate these systems using different sets of genes and on different temporal scales relative to the intensity of drought stress. We also find strong evidence that drought tolerant umbrella acacias demonstrate a surprisingly limited and relatively passive transcriptional response to drought stress, while splendid thorn acacias attempt to actively combat drought stress and maintain a steady state of growth and photosynthesis. Our study provides the first transcriptomic analysis of African acacias and a new model for investigating transcriptomic reactions over long periods of stress.
Keywords: Fabaceae; Vachellia robusta; Vachellia tortilis; acacia; differential gene expression; differential gene reaction; drought stress; savanna; transcriptome; water use efficiency.
© 2025 The Author(s). The Plant Journal published by Society for Experimental Biology and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Anderson, T.M. , Morrison, T. , Rugemalila, D. & Holdo, R. (2015) Compositional decoupling of savanna canopy and understory tree communities in Serengeti. Journal of Vegetation Science, 26, 385–394. Available from: 10.1111/jvs.12241 - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

