Rapid, interpretable data-driven models of neural dynamics using recurrent mechanistic models
- PMID: 40758876
- PMCID: PMC12358897
- DOI: 10.1073/pnas.2426916122
Rapid, interpretable data-driven models of neural dynamics using recurrent mechanistic models
Abstract
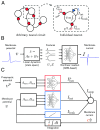
Obtaining predictive models of a neural system is notoriously challenging. Detailed models suffer from excess model complexity and are difficult to fit efficiently. Simplified models must negotiate a tradeoff between tractability, predictive power, and ease of interpretation. We present a modeling paradigm for estimating predictive, mechanistic models of neurons and small circuits that navigates these issues using methods from systems theory. The key insight is that membrane currents can be modeled using two scalable system components optimized for learning: linear state space models, and nonlinear artificial neural networks. Combining these components, we construct two types of membrane currents: lumped currents, which are flexible, and data-driven conductance-based currents, which are interpretable. The resulting class of models-which we call recurrent mechanistic models (RMMs)-can be trained in a matter of seconds to minutes on intracellular recordings during an electrophysiology experiment, representing a step change in performance over previous approaches. As a proof-of-principle, we use RMMs to learn the dynamics of two groups of neurons, and their synaptic connections, in the Stomatogastric Ganglion, a well-known central pattern generator. Due to their reliability, efficiency, and interpretability, RMMs enable qualitatively new kinds of experiments using predictive models in closed-loop neurophysiology and online estimation of neural properties in living preparations.
Keywords: biophysical models; central pattern generators; electrophysiology; machine learning; neural dynamics.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



References
-
- Paninski L., Pillow J., Simoncelli E., Comparing integrate-and-fire models estimated using intracellular and extracellular data. Neurocomputing 65, 379–385 (2005).
-
- Abarbanel H., Creveling D., Farsian R., Kostuk M., Dynamical state and parameter estimation. SIAM J. Appl. Dyn. Syst. 8, 1341–1381 (2009).
-
- Meliza C. D., et al. , Estimating parameters and predicting membrane voltages with conductance-based neuron models. Biol. Cybern. 108, 495–516 (2014). - PubMed
-
- J. M. Lueckmann et al. , “Flexible statistical inference for mechanistic models of neural dynamics” in 31st Conference on Neural Information Processing Systems (NIPS 2017), I. Guyon et al. , Eds. (Long Beach, CA, USA, 2017), pp. 1289–1299.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

