Detecting likely germline variants during tumor-based molecular profiling
- PMID: 40759574
- PMCID: PMC12321393
- DOI: 10.1172/JCI190264
Detecting likely germline variants during tumor-based molecular profiling
Abstract
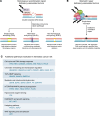
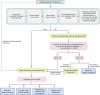
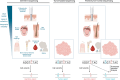
As the use of molecular profiling of tumors expands, cancer diagnosis, prognosis, and treatment planning increasingly rely on the information it provides. Although primarily designed to detect somatic variants, next-generation sequencing (NGS) tumor-based profiling also identifies germline DNA alterations, necessitating careful clinical interpretation of the data. Traditionally, germline risk testing has depended on prioritizing individuals based on physical exam findings consistent with known hereditary cancer syndromes, tumor-specific features, age at diagnosis, personal history, and family history. As NGS-based molecular profiling is used increasingly to diagnose, prognosticate, and follow cancer progression, DNA variants that are likely to be of germline origin are identified with increased frequency. Because pathogenic/likely pathogenic germline variants are critical biomarkers for risk stratification and treatment planning, consensus guidelines are expanding to recommend comprehensive germline testing for more cancer patients. This Review highlights the nuances of identifying DNA variants of potential germline origin incidentally at the time of NGS-based molecular profiling and emphasizes key differences between comprehensive germline versus tumor-based platforms, sample types, and analytical methodologies. In the growing era of precision oncology, clinicians should be adept at navigating these distinctions to optimize testing strategies and leverage insights regarding germline cancer risk surveillance and management for all people with cancer.
Conflict of interest statement
Figures
Similar articles
-
Can a Liquid Biopsy Detect Circulating Tumor DNA With Low-passage Whole-genome Sequencing in Patients With a Sarcoma? A Pilot Evaluation.Clin Orthop Relat Res. 2025 Jan 1;483(1):39-48. doi: 10.1097/CORR.0000000000003161. Epub 2024 Jun 21. Clin Orthop Relat Res. 2025. PMID: 38905450
-
Li-Fraumeni Syndrome.1999 Jan 19 [updated 2025 May 1]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. 1999 Jan 19 [updated 2025 May 1]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. PMID: 20301488 Free Books & Documents. Review.
-
Citrullinemia Type I.2004 Jul 7 [updated 2022 Aug 18]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. 2004 Jul 7 [updated 2022 Aug 18]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. PMID: 20301631 Free Books & Documents. Review.
-
Multidisciplinary collaborative guidance on the assessment and treatment of patients with Long COVID: A compendium statement.PM R. 2025 Jun;17(6):684-708. doi: 10.1002/pmrj.13397. Epub 2025 Apr 22. PM R. 2025. PMID: 40261198
-
Identification and triage of patients with incidental germline pathogenic variants on somatic tumor profiling with a genomics module.J Genet Couns. 2025 Jun;34(3):e70071. doi: 10.1002/jgc4.70071. J Genet Couns. 2025. PMID: 40537894
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

