Strengthening phage resistance of Streptococcus thermophilus by leveraging complementary defense systems
- PMID: 40759662
- PMCID: PMC12322248
- DOI: 10.1038/s41467-025-62408-3
Strengthening phage resistance of Streptococcus thermophilus by leveraging complementary defense systems
Abstract
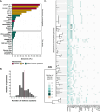
CRISPR-Cas and restriction-modification systems represent the core defense arsenal in Streptococcus thermophilus, but their effectiveness is compromised by phages encoding anti-CRISPR proteins (ACRs) and other counter-defense strategies. Here, we explore the defensome of 263 S. thermophilus strains to uncover other anti-phage systems. The defense landscape of S. thermophilus is enriched by 21 accessory defense systems, 13 of which have never been investigated in this species. Experimental validation of 17 systems with 14 phages reveals a range of anti-phage activities, highlighting both broad and narrow specificities across the five viral genera infecting S. thermophilus. Synergies are observed when combining CRISPR immunity with accessory systems. We also assess the fitness cost associated with the chromosomal integration of these systems in their native context and find no impact under laboratory or industrial conditions. These findings underscore the potential of these accessory defense systems to enhance the resistance of S. thermophilus, particularly against ACR-encoding phages.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: D.A.R., P.H., G.M.R. and S.M. are co-inventors on patent(s) or patent application(s) related to ACR and their various uses (US11732251, US11530405). The remaining authors declare no competing interests.
Figures




References
-
- Ortiz Charneco, G. et al. Bacteriophages in the dairy industry: A problem solved? Annu. Rev. Food Sci. Technol.14, 367–385 (2023). - PubMed
-
- Romero, D. A., Magill, D., Millen, A., Horvath, P. & Fremaux, C. Dairy lactococcal and streptococcal phage–host interactions: an industrial perspective in an evolving phage landscape. FEMS Microbiol. Rev.44, 909–932 (2020). - PubMed
-
- Mayo-Muñoz, D., Pinilla-Redondo, R., Birkholz, N. & Fineran, P. C. A host of armor: Prokaryotic immune strategies against mobile genetic elements. Cell Rep.42, 112672 (2023). - PubMed
-
- Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science315, 1709–1712 (2007). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

