Real-time capture of σN transcription initiation intermediates reveals mechanism of ATPase-driven activation by limited unfolding
- PMID: 40759887
- PMCID: PMC12322047
- DOI: 10.1038/s41467-025-61837-4
Real-time capture of σN transcription initiation intermediates reveals mechanism of ATPase-driven activation by limited unfolding
Abstract
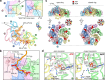
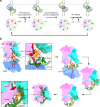
Bacterial σ factors bind RNA polymerase (E) to form holoenzyme (Eσ), conferring promoter specificity to E and playing a key role in transcription bubble formation. σN is unique among σ factors in its structure and functional mechanism, requiring activation by specialized AAA+ ATPases. EσN forms an inactive promoter complex where the N-terminal σN region I (σN-RI) threads through a small DNA bubble. On the opposite side of the DNA, the ATPase engages σN-RI within the pore of its hexameric ring. Here, we perform kinetics-guided structural analysis of de novo formed EσN initiation complexes and engineer a biochemical assay to measure ATPase-mediated σN-RI translocation during promoter melting. We show that the ATPase exerts mechanical action to translocate about 30 residues of σN-RI through the DNA bubble, disrupting inhibitory structures of σN to allow full transcription bubble formation. A local charge switch of σN-RI from positive to negative may help facilitate disengagement of the otherwise processive ATPase, allowing subsequent σN disentanglement from the DNA bubble.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures




Update of
-
Real-time capture of σN transcription initiation intermediates reveals mechanism of ATPase-driven activation by limited unfolding.bioRxiv [Preprint]. 2025 Feb 8:2025.02.07.637174. doi: 10.1101/2025.02.07.637174. bioRxiv. 2025. Update in: Nat Commun. 2025 Aug 4;16(1):7138. doi: 10.1038/s41467-025-61837-4. PMID: 39974980 Free PMC article. Updated. Preprint.
Similar articles
-
Real-time capture of σN transcription initiation intermediates reveals mechanism of ATPase-driven activation by limited unfolding.bioRxiv [Preprint]. 2025 Feb 8:2025.02.07.637174. doi: 10.1101/2025.02.07.637174. bioRxiv. 2025. Update in: Nat Commun. 2025 Aug 4;16(1):7138. doi: 10.1038/s41467-025-61837-4. PMID: 39974980 Free PMC article. Updated. Preprint.
-
Subunit specialization in AAA+ proteins and substrate unfolding during transcription complex remodeling.Proc Natl Acad Sci U S A. 2025 Apr 29;122(17):e2425868122. doi: 10.1073/pnas.2425868122. Epub 2025 Apr 24. Proc Natl Acad Sci U S A. 2025. PMID: 40273105 Free PMC article.
-
A general mechanism for transcription bubble nucleation in bacteria.Proc Natl Acad Sci U S A. 2023 Apr 4;120(14):e2220874120. doi: 10.1073/pnas.2220874120. Epub 2023 Mar 27. Proc Natl Acad Sci U S A. 2023. PMID: 36972428 Free PMC article.
-
Mechanisms of σ54-Dependent Transcription Initiation and Regulation.J Mol Biol. 2019 Sep 20;431(20):3960-3974. doi: 10.1016/j.jmb.2019.04.022. Epub 2019 Apr 25. J Mol Biol. 2019. PMID: 31029702 Free PMC article. Review.
-
Structure and molecular mechanism of bacterial transcription activation.Trends Microbiol. 2024 Apr;32(4):379-397. doi: 10.1016/j.tim.2023.10.001. Epub 2023 Oct 28. Trends Microbiol. 2024. PMID: 37903670 Review.
References
-
- Gruber, T. M. & Gross, C. A. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol.57, 441–466 (2003). - PubMed
-
- Correa, N. E., Lauriano, C. M., McGee, R. & Klose, K. E. Phosphorylation of the flagellar regulatory protein FlrC is necessary for Vibrio cholerae motility and enhanced colonization. Mol. Microbiol.35, 743–755 (2000). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

