Virus-human chromatin interactions reorganise 3D genome and hijack KDM5B for promoting metastasis in nasopharyngeal carcinoma
- PMID: 40759888
- PMCID: PMC12322282
- DOI: 10.1038/s41467-025-61597-1
Virus-human chromatin interactions reorganise 3D genome and hijack KDM5B for promoting metastasis in nasopharyngeal carcinoma
Abstract
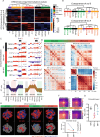
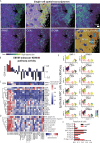
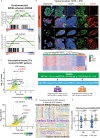
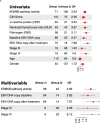
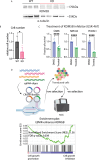
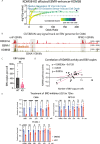
Epstein-Barr virus, the first identified human DNA tumour virus, is detectable in more than 90% of nasopharyngeal carcinoma patients in endemic regions. The 3D chromosome conformation analysis reveals that virus‒host chromatin interactions induce the spatial reorganisation of loops and compartments, resulting in "enhancer infestation" and switch of "H3K27 bivalency" at EBV-interacting regions. Through this mechanism, EBV hijacks KDM5B, a gatekeeper of genome stability, and its binding targets, leading to aberrant activation of an EBVIR-enhancer-KDM5B signature. Cancer cells with this signature present increased MYC activation, DNA damage responses, and epigenetic plasticity of epithelial-immune dual features with metastatic potential. Our multicentre multiomics study confirms that this signature is the prerequisite for chromosome instability and can be utilised as a risk factor for distant metastasis. This study highlights a mechanism in which latent viral episomes can alter the host high-order epigenotype, promoting transcriptional rewiring and metastasis in NPC.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
- 17102619/Research Grants Council, University Grants Committee (RGC, UGC)
- T12-703/22-R; T123-70323-N/Research Grants Council, University Grants Committee (RGC, UGC)
- 08192296/Food and Health Bureau (Food and Health Bureau of the Government of the Hong Kong Special Administrative Region)
- 2021A0505110010/Guangdong Science and Technology Department (Science and Technology Department, Guangdong Province)
- Health@InnoHK/Innovation and Technology Commission (ITF)
LinkOut - more resources
Full Text Sources

