Integrative metabolome and genome-wide transcriptome analyses reveal the regulatory network for bioactive compound biosynthesis in lettuce upon UV-A radiation
- PMID: 40759986
- PMCID: PMC12323076
- DOI: 10.1186/s43897-025-00163-1
Integrative metabolome and genome-wide transcriptome analyses reveal the regulatory network for bioactive compound biosynthesis in lettuce upon UV-A radiation
Abstract
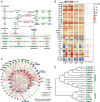
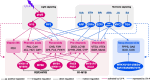
Ultraviolet A (UV-A) radiation possesses great potential for enhancing the bioactive properties of vegetables and also has promising application prospects in controlled-environment agriculture. Lettuce is a widely cultivated model vegetable in controlled-environment agriculture with abundant health-beneficial bioactive compounds. However, the comprehensive regulatory effectiveness and mechanism of UV-A on bioactive compounds in lettuce remain largely unclear. To address this issue, we performed transcriptomic and metabolomic analyses of UV-A-treated lettuce to construct a global map of metabolic features and transcriptional regulatory networks for all major bioactive compounds. Our study revealed that UV-A promotes the accumulation of most phenylpropanoids and vitamins (provitamin A and vitamin E/K1/B6) but represses the biosynthesis of sesquiterpenoids. MYB transcription factors (TFs) are key activators of bioactive compound biosynthesis promoted by UV-A, whereas WRKY TFs primarily inhibit the production of sesquiterpenoids. Moreover, light signaling plays a crucial and direct regulatory function in stimulating the biosynthesis of phenylpropanoids and vitamins but not in that of sesquiterpenoids. In comparison, hormone signaling dominates a more decisive regulatory role in repressing sesquiterpenoid biosynthesis through working directly and interacting with WRKY TFs. This study paves the way toward an understanding of the bioactive compound regulation and genetic improvement of lettuce bioactivity value.
Keywords: MYB; Phenylpropanoid; Sesquiterpenoid; Transcriptional regulation; Vitamin; WRKY.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: All authors approve the manuscript and consent to the publication of the work. Competing interests: The authors declare that they have no competing interests.
Figures






References
-
- Agati G, Tattini M. Multiple functional roles of flavonoids in photoprotection. New Phytol. 2010;186(4):786–93. - PubMed
-
- Agati G, Azzarello E, Pollastri S, Tattini M. Flavonoids as antioxidants in plants: location and functional significance. Plant Sci. 2012;196:67–76. - PubMed
-
- Anilakumar KR, Harsha SN. Mallesha, Sharma RK, Lettuce: a promising leafy vegetable with functional properties. Def Life Sci J. 2017;2(2):178–85.
-
- Asensi-Fabado MA, Munne-Bosch S. Vitamins in plants: occurrence, biosynthesis and antioxidant function. Trends Plant Sci. 2010;15(10):582–92. - PubMed
-
- Bhat ZY, Mir JA, Yadav AK, Singh D, Ashraf N. CstMYB1R1, a REVEILLE-8-like transcription factor, regulates diurnal clock-specific anthocyanin biosynthesis and response to abiotic stress in Crocus sativus L. Plant Cell Rep. 2024;43(1):20. - PubMed
Grants and funding
- 32171890/National Natural Science Foundation of China
- 32401686/National Natural Science Foundation of China
- 2024(2)/Earmarked fund for Shanghai Modern Leaf-vegetable industry Technology Research System
- 23N21900300/Science and Technology Commission of Shanghai Municipality
- 2021M702130/China Postdoctoral Science Foundation
LinkOut - more resources
Full Text Sources
