Uncovering diversity and abundance patterns of CO2-fixing microorganisms in peatlands
- PMID: 40760046
- PMCID: PMC12322008
- DOI: 10.1038/s44185-025-00099-1
Uncovering diversity and abundance patterns of CO2-fixing microorganisms in peatlands
Abstract
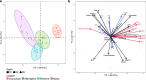
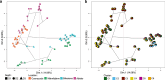
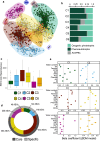
Microorganisms play a crucial role in the carbon (C) dynamics of peatlands - a major terrestrial C reservoir. Because of their role in C emissions, heterotrophic microorganisms have attracted much attention over the past decades. CO2-fixing microorganisms (CFMs) remained largely overlooked, while they could attenuate C emissions. Here, we use metabarcoding and digital droplet PCR to survey microorganisms that potentially fix CO2 in different peatlands. We demonstrate that CFMs are abundant and diverse in peatlands, with on average 1021 CFMs contributing up to 40% of the total bacterial abundance. Using a joint-species distribution model, we identified a core and a specific CFM microbiome, the latter being influenced by temperature and nutrients. Our findings highlight that ASV richness and community structure were direct drivers of CFM abundance, while environmental parameters were indirect drivers. These results provide the basis for a better understanding of the role of CFMs in peatland C cycle inputs.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Nichols, J. E. & Peteet, D. M. Rapid expansion of northern peatlands and doubled estimate of carbon storage. Nat. Geosci.12, 917–921 (2019).
-
- Rydin, H. & Jeglum, J. K. The biology of peatlands. 10.1093/ACPROF:OSO/9780198528722.001.0001 (2006).
-
- Strack, M., Davidson, S. J., Hirano, T. & Dunn, C. The potential of peatlands as nature-based climate solutions. Curr. Clim. Change Rep.8, 71–82 (2022).
-
- Hamard, S., Küttim, M., Céréghino, R. & Jassey, V. E. J. Peatland microhabitat heterogeneity drives phototrophic microbe distribution and photosynthetic activity. Environ. Microbiol.23, 6811–6827 (2021). - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

