Uncovering biomarkers and pathways in oral squamous cell carcinoma through integrated lncRNA-mRNA regulatory network analysis
- PMID: 40760230
- PMCID: PMC12321711
- DOI: 10.1007/s12672-025-02922-4
Uncovering biomarkers and pathways in oral squamous cell carcinoma through integrated lncRNA-mRNA regulatory network analysis
Abstract
Objective: This study aimed to identify potential biomarkers and elucidate molecular mechanisms in oral squamous cell carcinoma (OSCC) by leveraging an integrated bioinformatics approach.
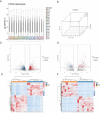
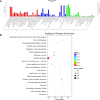
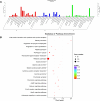
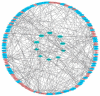
Methods: We conducted high-throughput RNA sequencing on OSCC and adjacent normal tissues to profile mRNA and lncRNA expression. Differentially expressed genes were identified and subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. A competing endogenous RNA (ceRNA) network was constructed, and protein-protein interaction (PPI) networks were analyzed using STRING and Cytoscape software. Hub genes were identified using the Cytohubba plug-in in Cytoscape.
Results: Our analysis identified 5362 differentially expressed mRNAs and 2801 differentially expressed lncRNAs. GO analysis revealed that dysregulated mRNAs were associated with system development and responses to organic substances. At the same time, lncRNAs were enriched in muscle system processes and cell-cell signaling. KEGG pathway analysis highlighted cancer-related pathways, including cytokine-cytokine receptor interactions and the NF-kappa B signaling pathway. The constructed ceRNA network highlighted key regulatory nodes, including hub genes IGF2BP1, CLDN6, and HLA-G, which may play pivotal roles in OSCC.
Conclusions: This study provides a comprehensive lncRNA-mRNA regulatory network and identifies biomarkers that could advance OSCC therapeutic strategies. The findings offer new insights into OSCC pathogenesis and potential targets for clinical application.
Keywords: Bioinformatics; CeRNA network; LncRNA; Oral squamous cell carcinoma; mRNA.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Consent for publication: Informed consent was obtained from all individual participants included in the study. Ethical approval: All procedures performed in studies involving human participants were in accordance with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. This study is approved by the Ethics Committee of Foshan First People’s Hospital. Written informed consent was obtained. Competing interests: The authors declare no competing interests.
Figures
Similar articles
-
Identifying pyroptosis- and inflammation-related genes in spinal cord injury based on bioinformatics analysis.Sci Rep. 2025 Jul 14;15(1):25424. doi: 10.1038/s41598-025-10541-w. Sci Rep. 2025. PMID: 40659785 Free PMC article.
-
The mechanism of arsenic trioxide and microwave ablation in the treatment of oral squamous cell carcinoma based on high throughput sequencing.IET Syst Biol. 2025 Jan-Dec;19(1):e12113. doi: 10.1049/syb2.12113. Epub 2024 Dec 23. IET Syst Biol. 2025. PMID: 39716349 Free PMC article.
-
Bioinformatics identification and validation of m6A/m1A/m5C/m7G/ac4 C-modified genes in oral squamous cell carcinoma.BMC Cancer. 2025 Jul 1;25(1):1055. doi: 10.1186/s12885-025-14216-7. BMC Cancer. 2025. PMID: 40597017 Free PMC article.
-
Medicine for chronic atrophic gastritis: a systematic review, meta- and network pharmacology analysis.Ann Med. 2023;55(2):2299352. doi: 10.1080/07853890.2023.2299352. Epub 2024 Jan 3. Ann Med. 2023. PMID: 38170849 Free PMC article.
-
Deciphering the miRNA-mRNA Interaction Network Regulating Aging Skeletal Muscle in Various Exercise Regimens through Comprehensive Bioinformatics Analysis.Cell Biochem Biophys. 2025 Aug 1. doi: 10.1007/s12013-025-01848-6. Online ahead of print. Cell Biochem Biophys. 2025. PMID: 40748585 Review.
References
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. 10.3322/caac.21660. - PubMed
-
- Weckx A, Grochau KJ, Grandoch A, Backhaus T, Zoller JE, Kreppel M. Survival outcomes after surgical treatment of oral squamous cell carcinoma. Oral Dis. 2020;26(7):1432–9. 10.1111/odi.13422. - PubMed
-
- Dik EA, Willems SM, Ipenburg NA, Adriaansens SO, Rosenberg AJ, van Es RJ. Resection of early oral squamous cell carcinoma with positive or close margins: relevance of adjuvant treatment in relation to local recurrence: margins of 3 mm as safe as 5 mm. Oral Oncol. 2014;50(6):611–5. 10.1016/j.oraloncology.2014.02.014. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
