A programmed decline in ribosome levels governs human early neurodevelopment
- PMID: 40760247
- PMCID: PMC12339376
- DOI: 10.1038/s41556-025-01708-8
A programmed decline in ribosome levels governs human early neurodevelopment
Abstract
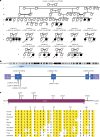
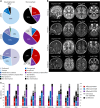
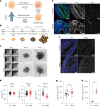
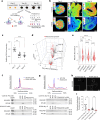
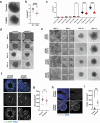
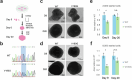
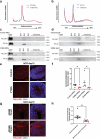
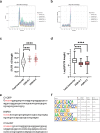
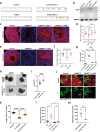
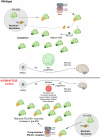
Many neurodevelopmental defects are linked to genes involved in housekeeping functions, such as those encoding ribosome biogenesis factors. How reductions in ribosome biogenesis can result in tissue- and developmental-specific defects remains unclear. Here we describe variants in the ribosome biogenesis factor AIRIM/C1orf109 that are primarily associated with neurodevelopmental disorders. Using human cerebral organoids in combination with proteomic, single-cell RNA sequencing and single-organoid translation analyses, we identify a previously unappreciated drop in protein production during early brain development. We find that ribosome levels decrease during neuroepithelial differentiation, making differentiating cells particularly vulnerable to perturbations in ribosome biogenesis during this time. Reduced ribosome availability more profoundly impacts the translation of specific transcripts, disrupting both survival and cell fate commitment of transitioning neuroepithelia. Enhancing mTOR activity suppresses the growth and developmental defects associated with AIRIM/C1orf109 variants. This work provides evidence for the functional importance of regulated changes in global protein synthesis capacity during cellular differentiation.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures



References
-
- Parenti, I., Rabaneda, L. G., Schoen, H. & Novarino, G. Neurodevelopmental disorders: from genetics to functional pathways. Trends Neurosci.43, 608–621 (2020). - PubMed
MeSH terms
Substances
Grants and funding
- XDB0820000/Chinese Academy of Sciences (CAS)
- R01 GM101149/GM/NIGMS NIH HHS/United States
- F-2027-20230405/Welch Foundation
- GM101149/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R35GM144043/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- I-2088/Welch Foundation
- DFG VO 2138/7-1/Deutsche Stiftung Friedensforschung (German Foundation for Peace Research)
- 222531/Z/21/Z/Wellcome Trust (Wellcome)
- R01 GM138565/GM/NIGMS NIH HHS/United States
- RR180042/Cancer Prevention and Research Institute of Texas (Cancer Prevention Research Institute of Texas)
- R01AG079513/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- BB/Z51522X/1/RCUK | Biotechnology and Biological Sciences Research Council (BBSRC)
- F31 NS125906/NS/NINDS NIH HHS/United States
- R35 GM144043/GM/NIGMS NIH HHS/United States
- NS125906/U.S. Department of Health & Human Services | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R01AG067604/U.S. Department of Health & Human Services | NIH | National Institute on Aging (U.S. National Institute on Aging)
- R01 AG079513/AG/NIA NIH HHS/United States
- HD103627/U.S. Department of Health & Human Services | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- WT_/Wellcome Trust/United Kingdom
- CA174905/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- GM138565/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R01 HD103627/HD/NICHD NIH HHS/United States
- R01 AG067604/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

