Bladder cancer microbiome and its association with chemoresponse
- PMID: 40761254
- PMCID: PMC12318727
- DOI: 10.3389/fonc.2025.1506319
Bladder cancer microbiome and its association with chemoresponse
Abstract
Background: The microbiome is widely known to cause come types of cancer, modify cancer biology, and impact therapeutic efficacy. Despite the urinary microbiome being one of the most clinically significant microbiomes in human health, it is one of the least well-described.
Methods and materials: To begin to annotate the urinary microbiome present in bladder cancers, we analyzed human genome-filtered sequencing data from the Cancer Genome Atlas (TCGA) from 116 tumors (duplicates from 22 tumors), 22 adjacent normal bladder tissues, and 99 blood samples to classify reads originating from known microbiota. We also performed 16S rRNA sequencing on urine samples from 55 patients with urothelial carcinoma and 13 non-cancer patients. Additionally, we compared microbiome from matched urine samples and archival diagnostic tumor samples from 21 bladder cancer patients. For our animal experiments, starting at 8-12 weeks of age, male (n=33) and female (n=22) C57BL/6 mice were administered 0.05% N-butyl-N-(4-hydroxybutyl)-nitrosamine (BBN) in drinking water for 12 weeks then switched to regular water. Control mice drank regular water. Bladders were collected for 16S rRNA sequencing pre-exposure, after 6 and 12 weeks of exposure, and at the time of tumor identification (typically between 14-22 weeks from start of treatment using this model). Finally, because of our findings, we tested the effects of E.coli infection on gemcitabine toxicity in bladder cancer cell lines.
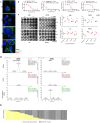
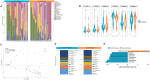
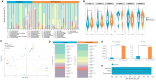
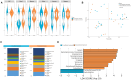
Results: Twenty-seven viral and bacterial species were found to be enriched in the tumor samples from TCGA cohort, including sex-specific enrichment of Lactobacillus and Prevotella in female bladder cancer patients which also are prevalent in the normal female genitourinary tract. We found key differences in urinary microbiota profiles that distinguish cancer patients from healthy control. We also found Granulicatella and Proteus were enriched in patients who did not respond to neoadjuvant chemotherapy while E. Faecalis was enriched in responders. Additionally, we found 32% overlap between microbiota of urine and archival diagnostic tumor samples. Because bladder cancer patients undergoing surgical procedures are exposed to a single dose of peri-procedure antibiotics, we took advantage of a 'natural experiment' to measure microbial changes in urology patients receiving a single dose of antibiotics for skin procedures, finding that there are very few changes that persist for 1 month. Additionally, we measured microbial changes during BBN-induced carcinogenesis in a mouse model and observed that changes are more likely related to BBN exposure itself rather than carcinogenesis as changes induced by BBN resolve after BBN withdrawal. Lastly, urine from bladder cancer patients harbored abundant Gammaproteobacteria, which in cell culture experiments, detoxified gemcitabine, a commonly used therapy in bladder cancer.
Conclusions: In summary, we identify many new relationships between the microbiome and bladder cancer that are clinically relevant and lay the groundwork important functional studies in the future.
Keywords: 16S sequencing; BBN; bladder cancer; gemcitabine; microbiome; microbiota; urothelial carcinoma.
Copyright © 2025 Ginwala, Bukavina, Sindhani, Nachman, Peri, Franklin, Drevik, Christianson, Geynisman, Kutikov and Abbosh.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2020 Jan 9;1(1):CD011535. doi: 10.1002/14651858.CD011535.pub3. Cochrane Database Syst Rev. 2020. Update in: Cochrane Database Syst Rev. 2021 Apr 19;4:CD011535. doi: 10.1002/14651858.CD011535.pub4. PMID: 31917873 Free PMC article. Updated.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
A New Measure of Quantified Social Health Is Associated With Levels of Discomfort, Capability, and Mental and General Health Among Patients Seeking Musculoskeletal Specialty Care.Clin Orthop Relat Res. 2025 Apr 1;483(4):647-663. doi: 10.1097/CORR.0000000000003394. Epub 2025 Feb 5. Clin Orthop Relat Res. 2025. PMID: 39915110
References
-
- Rajagopala SV, Vashee S, Oldfield LM, Suzuki Y, Venter JC, Telenti A, et al. The Human Microbiome and Cancer. Cancer Prev Res (Phila). (2017) 10:226–34. - PubMed
-
- Bukavina L, Isali I, Ginwala R, Sindhani M, Calaway A, Magee D, et al. Global Meta-analysis of Urine Microbiome: Colonization of Polycyclic Aromatic Hydrocarbon-degrading Bacteria Among Bladder Cancer Patients. Eur Urol oncology. (2023) 6:190–203. doi: 10.1016/j.euo.2023.02.004, PMID: - DOI - PubMed
-
- Fouts DE, Pieper R, Szpakowski S, Pohl H, Knoblach S, Suh MJ, et al. Integrated next-generation sequencing of 16S rDNA and metaproteomics differentiate the healthy urine microbiome from asymptomatic bacteriuria in neuropathic bladder associated with spinal cord injury. J Trans medicine. (2012) 10:174. doi: 10.1186/1479-5876-10-174, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

