Molecular characteristics and antimicrobial susceptibility of carbapenem-resistant Klebsiella pneumoniae in a multicenter study in Ningbo, China
- PMID: 40761276
- PMCID: PMC12318958
- DOI: 10.3389/fmicb.2025.1628592
Molecular characteristics and antimicrobial susceptibility of carbapenem-resistant Klebsiella pneumoniae in a multicenter study in Ningbo, China
Abstract
Objective: To analyze the molecular epidemiology and antimicrobial resistance profiles of carbapenem-resistant Klebsiella pneumoniae (CR-KP) isolates in Ningbo, with the aim of providing a theoretical basis for hospital infection control strategies and the implementation of precise clinical diagnosis and treatment protocols.
Methods: During the period from April 30, 2023 to June 30, 2024, clinical isolates of Klebsiella pneumoniae were collected from multiple centers in Ningbo, including The Affiliated Li Huili Hospital (Yinzhou District, Ningbo), Xiangshan Red Cross Taiwan Compatriot Hospital Medical and Health Group (Xiangshan, Ningbo), and the Second Hospital of Ninghai County (Ninghai, Ningbo). A total of 81 CR-KP strains were identified using the broth dilution method for carbapenem resistance screening. These isolates were submitted to Beijing Novo gene Co., Ltd. for sequencing analysis. The sequencing data were analyzed using online tools (https://bigsdb.pasteur.fr/ and http://genepi.food.dtu.dk/resfinder) to obtain information on multilocus sequence typing (MLST), capsular serotype (KL type), virulence genes, and resistance genes. Phylogenetic relationships were constructed using SNP software. For plasmid characterization, the PlasmidFinder online tool (https://cge.food.dtu.dk/services/PlasmidFinder/) was utilized to identify plasmid replicon genes and perform Inc. typing analysis. Furthermore, to conduct a comprehensive collinearity analysis of the bla KPC-2 resistance plasmid gene, gene cluster maps were constructed using Bakta v1.11.0 and Clinker v0.0.28 software packages.
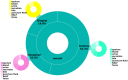
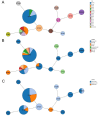
Results: Among the 81 CR-KP isolates, MLST typing revealed that ST11 was the predominant sequence type, accounting for 66.67% (54/81), with KL64 being the dominant capsular type. Among the non-ST11 CR-KP isolates, the ST15 type accounted for 48.15% (13/27), with KL19 being the predominant capsular serotype. The carriage rate of virulence genes-including rmpA2, fyuA, and 10 other genes-was significantly higher in ST11 CR-KP compared to non-ST11 CR-KP (p < 0.05). Analysis of resistance genes revealed that ST11 CR-KP primarily carried bla KPC-2 (100%, 54/54), whereas the resistance gene profiles among non-ST11 CR-KP isolates were more diverse, including blaNDM , bla IMP, and bla OXA. Plasmid typing indicated that ST11 CR-KP predominantly harbored IncFII (98.15%, 53/54) and RepB (72.22%, 39/54) plasmid types. In contrast, non-ST11 CR-KP isolates exhibited a wider range of plasmid types, including IncX3 (33.33%, 9/27), RepB (25.93%, 7/27), IncFII (25.93%, 7/27), IncFIB (7.41%, 2/27), and both ColKP3 and Col440II (7.41%, 2/27). Antimicrobial susceptibility testing demonstrated high resistance rates to commonly used antibiotics in both ST11 and non-ST11 CR-KP isolates. ST11 CR-KP exhibited 100% resistance to six antibiotics, including ceftriaxone (CRO), cefotetan (CTT), and cefepime (FEP), and showed susceptibility only to gentamicin (GEN), aztreonam/avibactam (AZA), ceftazidime/avibactam (CZA), polymyxin B (POL), and tigecycline (TGC). Non-ST11 CR-KP showed a significantly higher resistance rate to gentamicin (GEN) and ceftazidime/avibactam (CZA) than ST11 CR-KP (p < 0.05), but lower resistance rates to cefotetan (74.07%), all of which were statistically significant (p < 0.05).
Conclusion: In the Ningbo region, CR-KP is predominantly of the ST11-KL64 type, exhibiting both strong antimicrobial resistance and high virulence characteristics. Non-ST11 CR-KP isolates carry genetically diverse carbapenemase genes and mobile genetic elements (e.g., IncX3, ColKP3). ST11 CR-KP strains demonstrate significantly stronger resistance profiles compared to non-ST11 strains. Therefore, stringent control over the use of carbapenem antibiotics is essential, along with measures to prevent the spread of resistance plasmids and the continuous improvement of hospital infection control strategies.
Keywords: Klebsiella pneumoniae; carbapenem resistance; plasmids introduction; resistance genes; virulence genes.
Copyright © 2025 Shi, Tu, Li, Gao, Wang, Zhang, Jiang, Sun, Bao, Yang and Chang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
Emergence of ST11-KL64 carbapenem-resistant hypervirulent Klebsiella Pneumoniae isolates harboring blaKPC-2 and iucA from a tertiary teaching hospital in Western China.BMC Infect Dis. 2025 Jul 1;25(1):880. doi: 10.1186/s12879-025-11241-6. BMC Infect Dis. 2025. PMID: 40596916 Free PMC article.
-
Antimicrobial resistance and phylogenetic lineages of KPC-2-producing blood-borne Klebsiella pneumoniae subsp. pneumoniae from Kolkata, India during 2015-2024: Emergence of Klebsiella pneumoniae subsp. pneumoniae with blaKPC-2, blaNDM, and blaOXA-48-like triple carbapenemases.Microbiol Spectr. 2025 Aug 5;13(8):e0012625. doi: 10.1128/spectrum.00126-25. Epub 2025 Jun 12. Microbiol Spectr. 2025. PMID: 40503825 Free PMC article.
-
High genome diversity of Klebsiella pneumoniae strains isolated from a Chinese traditional medicine hospital in Jiangsu province, China, from 2023 to 2024.Front Microbiol. 2025 Jul 9;16:1575216. doi: 10.3389/fmicb.2025.1575216. eCollection 2025. Front Microbiol. 2025. PMID: 40703230 Free PMC article.
-
Adefovir dipivoxil and pegylated interferon alfa-2a for the treatment of chronic hepatitis B: a systematic review and economic evaluation.Health Technol Assess. 2006 Aug;10(28):iii-iv, xi-xiv, 1-183. doi: 10.3310/hta10280. Health Technol Assess. 2006. PMID: 16904047
-
Home treatment for mental health problems: a systematic review.Health Technol Assess. 2001;5(15):1-139. doi: 10.3310/hta5150. Health Technol Assess. 2001. PMID: 11532236
References
LinkOut - more resources
Full Text Sources
Miscellaneous

