Distribution of human-pathogenic Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in crab-eating macaques in China
- PMID: 40761285
- PMCID: PMC12318935
- DOI: 10.3389/fmicb.2025.1641632
Distribution of human-pathogenic Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in crab-eating macaques in China
Abstract
Introduction: The positive rates and genetic identity of Cryptosporidium spp., Giardia duodenalis (G. duodenalis), and Enterocytozoon bieneusi (E. bieneusi) were unclear in crab-eating macaques in Suzhou and Beijing, China.
Methods: A total of 504 fecal samples were collected from crab-eating macaques on commercial farms in Beijing and Suzhou, China. The extracted DNA was analyzed for Cryptosporidium spp. and E. bieneusi by nested PCR and sequence analysis of the small subunit rRNA (SSU rRNA) gene and the internal transcribed spacer (ITS) gene, respectively. The G. duodenalis was detected by nested PCR targeting β-giardin (bg) gene, glutamate dehydrogenase (gdh) gene, and triosephosphate isomerase (tpi) gene. The C. hominis identified were further subtyped by nested PCR and sequence analysis of the 60 kDa glycoprotein (gp60) gene.
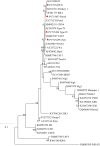
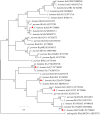
Results: All 504 fecal samples collected from crab-eating macaques, the detection rates of Cryptosporidium spp., G. duodenalis, and E. bieneusi were 11.9% (60/504), 5.6% (28/504), and 4.6% (23/504), respectively. The 15.1% (44/292) detection rate of Cryptosporidium spp. from crab-eating macaques in Suzhou was significantly higher than that in Beijing (2.8%; 6/212; χ 2 = 20.6, df = 1, p < 0.0001). The detection rates of Cryptosporidium spp. and G. duodenalis were significant different between <2 months old animals and >24 months old animals (χ 2 = 104.7, df = 1, p < 0.0001; χ 2 = 6.6, df = 1, p = 0.0104). In contrast, there was no significant different in the detection rate of E. bieneusi in two age groups (χ 2 = 2.2, df = 1, p = 0.1360). A total of one Cryptosporidium species, one G. duodenalis assemblage B, and 4 E. bieneusi genotypes have been identified, including C. hominis (n = 60), assemblage B (n = 28), CM1 (n = 14), Peru8 (n = 5), D (n = 3), and Type IV (n = 1). Among 60 C. hominis samples, five subtypes of five subtype families were successfully identified at the gp60 gene: IbA13G4 (n = 27), InA26 (n = 3), IfA17G2R3 (n = 3), IiA17 (n = 3), and IeA11G3T3 (n = 2).
Discussion: The results indicate that known zoonotic Cryptosporidium spp., G. duodenalis, and E. bieneusi are prevalent in crab-eating macaques. The crab-eating macaques could play a potential role in the zoonotic transmission of pathogens to humans.
Keywords: China; Cryptosporidium spp.; Enterocytozoon bieneusi; Giardia duodenalis; crab-eating macaque; zoonosis.
Copyright © 2025 Zhang, Chen, He, Li and Li.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources
Miscellaneous

