In vitro metabolic interaction network of a rationally designed nasal microbiota community
- PMID: 40761294
- PMCID: PMC12320084
- DOI: 10.1016/j.isci.2025.113114
In vitro metabolic interaction network of a rationally designed nasal microbiota community
Abstract
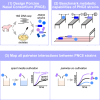
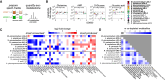
Mounting evidence suggests that metabolite exchange between microbiota members is a key driver of microbiota composition. However, we still know little about the metabolic interaction networks within many microbiota. To tackle this issue, we developed the porcine nasal consortium (PNC8), which represents the most in vivo abundant genera in the nasal microbiota of healthy piglets, and used it to systematically map the in vitro metabolic interactions between its members. Spent media experiments, exometabolomics, and direct co-cultivation, revealed that most pairwise interactions between PNC8 strains are negative, with co-depletion of sugars acting as a key driver. This prevalence of negative interactions leads to a complex competition hierarchy in which only few strains are able to consistently outcompete all others. Overall, this work provides a valuable resource for studying the nasal microbiota under experimentally tractable in vitro conditions and is a key step toward mapping its metabolic interaction network.
Keywords: metabolomics; microbiome.
© 2025 The Authors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Drugs for preventing postoperative nausea and vomiting in adults after general anaesthesia: a network meta-analysis.Cochrane Database Syst Rev. 2020 Oct 19;10(10):CD012859. doi: 10.1002/14651858.CD012859.pub2. Cochrane Database Syst Rev. 2020. PMID: 33075160 Free PMC article.
-
The effect of sample site and collection procedure on identification of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2024 Dec 16;12(12):CD014780. doi: 10.1002/14651858.CD014780. Cochrane Database Syst Rev. 2024. PMID: 39679851 Free PMC article.
-
Sexual Harassment and Prevention Training.2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2024 Mar 29. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 36508513 Free Books & Documents.
-
Community views on mass drug administration for soil-transmitted helminths: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2025 Jun 20;6(6):CD015794. doi: 10.1002/14651858.CD015794.pub2. Cochrane Database Syst Rev. 2025. PMID: 40539472 Review.
-
Incentives for preventing smoking in children and adolescents.Cochrane Database Syst Rev. 2017 Jun 6;6(6):CD008645. doi: 10.1002/14651858.CD008645.pub3. Cochrane Database Syst Rev. 2017. PMID: 28585288 Free PMC article.
References
-
- Osbelt L., Wende M., Almási É., Derksen E., Muthukumarasamy U., Lesker T.R., Galvez E.J.C., Pils M.C., Schalk E., Chhatwal P., et al. Klebsiella oxytoca causes colonization resistance against multidrug-resistant K. pneumoniae in the gut via cooperative carbohydrate competition. Cell Host Microbe. 2021;29:1663–1679.e7. - PubMed
-
- Eberl C., Weiss A.S., Jochum L.M., Durai Raj A.C., Ring D., Hussain S., Herp S., Meng C., Kleigrewe K., Gigl M., et al. E. coli enhance colonization resistance against Salmonella Typhimurium by competing for galactitol, a context-dependent limiting carbon source. Cell Host Microbe. 2021;29:1680–1692.e7. - PubMed
LinkOut - more resources
Full Text Sources

