Nanoparticle-Delivered siRNA Targeting NSUN4 Relieves Systemic Lupus Erythematosus through Declining Mitophagy-Mediated CD8+T Cell Exhaustion
- PMID: 40761479
- PMCID: PMC12318824
- DOI: 10.1002/mco2.70311
Nanoparticle-Delivered siRNA Targeting NSUN4 Relieves Systemic Lupus Erythematosus through Declining Mitophagy-Mediated CD8+T Cell Exhaustion
Abstract
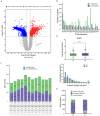
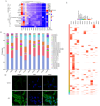
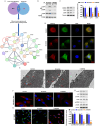
5-Methylcytosine modification (m5C) is an important posttranscriptional regulatory mechanism of gene expression. Exhausted CD8+T cells contribute to the development of many major diseases; however, their exact role and relationship to m5C in systemic lupus erythematosus (SLE) remain unknown. In this study, we identified a CD7highCD74high CD8+T subgroup that were robustly expanded in SLE patients through single-cell transcriptome sequencing (scRNA-seq). CD7highCD74high CD8+T cells displayed exhausted features and exhibited a superior diagnostic value in SLE. Then, we explored the m5C landscape of SLE patients by performing m5C epitranscriptome sequencing (m5C-seq). ScRNA-seq and m5C-seq were conjointly analyzed to screen m5C-related therapeutic targets for SLE, and NOP2/Sun RNA methyltransferase 4 (NSUN4) was identified as a key regulator of SLE pathogenesis. Knockdown of NSUN4 downregulated CD74 expression via reduction of m5C and suppressed CD8+T cell exhaustion by declining CD44/mTOR (mechanistic target of rapamycin kinase)-mediated mitophagy. Finally, we verified that nanoparticle-delivered siRNA against Nusn4 decreased autoimmune reaction kidney damage in both spontaneous and pristane-induced SLE mouse models. In conclusion, we identify an exhausted CD7highCD74high CD8+T cell subset and propose the crucial role of NSUN4/CD74-induced dysregulation of mitophagy in SLE pathogenesis, and targeting NSUN4 is a promising treatment strategy for SLE patients.
Keywords: CD8+T cell mitophagy and exhaustion; NSUN4; nanoparticle‐delivered siRNA; single‐cell RNA sequencing and m5C sequencing; systemic lupus erythematosus.
© 2025 The Author(s). MedComm published by Sichuan International Medical Exchange & Promotion Association (SCIMEA) and John Wiley & Sons Australia, Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




References
-
- Kiriakidou M., Ching C. L., “Systemic Lupus Erythematosus,” Annals of Internal Medicine 172, no. 11 (2020): 81–96. - PubMed
-
- Quintero‐González D. C., Muñoz‐Urbano M., Vásquez G. “Mitochondria as a Key Player in Systemic Lupus Erythematosus,” Autoimmunity 55, no. 8 (2022): 497–505. - PubMed
-
- Bolouri N., Akhtari M., Farhadi E., et al., “Role of the Innate and Adaptive Immune Responses in the Pathogenesis of Systemic Lupus Erythematosus,” Inflammation Research 71, no. 5‐6 (2022): 537–554. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
