Exploring the predictive "psycho-biomarkers" for checkpoint immunotherapy in cancer
- PMID: 40761787
- PMCID: PMC12318748
- DOI: 10.3389/fimmu.2025.1590670
Exploring the predictive "psycho-biomarkers" for checkpoint immunotherapy in cancer
Abstract
Background: In recent decades, cancer immunotherapy has transformed the treatment landscape, offering significant advantages over traditional therapies by improving progression-free survival (PFS) and overall survival (OS). However, immune checkpoint inhibitors (ICIs) treatment has been associated with an increased risk of mortality in its early stages. Therefore, identifying reliable biomarkers to predict which patients will benefit clinically from ICIs therapy is critical. Depression, a common form of chronic psychological stress, has emerged as a regulator of tumor immunity and is gaining attention as a target for novel cancer treatments. To date, no studies have explored the potential of depression-related genes in predicting response to ICIs therapy.
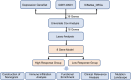
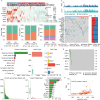
Methods: Public datasets of ICIs-treated patients were obtained from the TCGA and GEO databases, followed by comprehensive analyses, including bulk mRNA sequencing (mRNA-seq), co-expression network construction, and Gene Ontology enrichment. Regression analysis, using Cox proportional hazards and least absolute shrinkage and selection operator (Lasso), identified eight depression-related genes to build a predictive model for clinical outcomes in ICIs therapy. Additionally, correlations were explored between the depression-related predictive score and clinical parameters, including tumor mutational burden (TMB) and immune cell infiltration, establishing the score as a potential predictor of ICIs response.
Results: The model categorized patients into high- and low-responsiveness groups, with significant differences in disease-free survival (DFS) between them. Validation using both internal and external datasets demonstrated the model's strong predictive accuracy. Further analysis revealed that this response stratification correlates with immune cell abundance and TMB in cancer patients.
Conclusion: This study suggests that depression-related genetic traits could serve as biomarkers for ICIs therapy response, tumor mutations, and immune system alterations. Our findings offer insights into personalized therapeutic strategies for early intervention and prognosis in specific cancer types.
Keywords: breast cancer; immunotherapy efficacy; predictive model; psycho-biomarkers; psychological stress.
Copyright © 2025 Zuo, Chen, Xiao, Dai, Chen, Liang, Wu, Huang, Cui, Xu and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

