Differential DNA methylation patterns in whole blood from ACPA-positive patients with DMARD-naïve rheumatoid arthritis at clinical disease onset
- PMID: 40761796
- PMCID: PMC12318994
- DOI: 10.3389/fimmu.2025.1488161
Differential DNA methylation patterns in whole blood from ACPA-positive patients with DMARD-naïve rheumatoid arthritis at clinical disease onset
Abstract
Objective: Epigenetic DNA imprints are increasingly being recognized as co-drivers of disease in complex conditions. In this exploratory and hypothesis-generating epigenome-wide association study (EWAS), we investigated differential methylation patterns in peripheral blood leucocytes from patients with early untreated ACPA-positive rheumatoid arthritis (RA) versus controls.
Methods: Whole blood DNA was isolated from 101 disease-modifying anti-rheumatic drug (DMARD)-naïve patients with recent clinical onset of ACPA-positive RA and 200 controls. DNA methylation was studied using the Illumina MethylationEPIC BeadChips (Illumina). We assessed our findings against previously reported differentially methylated DNA positions associated with RA including an EWAS on peripheral blood leucocytes from a similar Drop Nordic cohort.
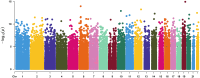
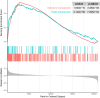
Results: We identified 16,583 CpG sites and 14 differentially methylated regions (DMRs) associated with RA. The most robust DMRs were in the gene body of LAMP1 and the TNSF14 GENE known as LIGHT. We identified three novel Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, the taste transduction pathway, the olfactory pathway, and the viral carcinogenesis pathway, which have not previously been associated with RA. We replicated 2,248 CpG sites reported earlier in an EWAS on peripheral blood leukocytes from RA patients of Scandinavian ancestry with incipient untreated ACPA-positive disease.
Conclusion: We have detected a considerable number of epigenetic marks with potential relevance to the pathogenesis of RA. These findings may pave the way for the development of narrowly targeted new drugs and possibly assist to retrieve persons at particular risk of acquiring RA.
Keywords: DNA-methylation; anti-CCP antibodies; epigenetics; incidence; rheumatoid arthritis.
Copyright © 2025 Svendsen, Mengel-From, Junker, Dalgård, Davey Smith, Relton, Elliott, Kyvik, Lindegaard, Christensen and Tan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Expansion of HLA-DR Positive Peripheral Helper T and Naive B Cells in Anticitrullinated Protein Antibody-Positive Individuals At Risk for Rheumatoid Arthritis.Arthritis Rheumatol. 2024 Jul;76(7):1023-1035. doi: 10.1002/art.42839. Epub 2024 Apr 6. Arthritis Rheumatol. 2024. PMID: 38412870 Free PMC article.
-
A systematic review of the effectiveness of adalimumab, etanercept and infliximab for the treatment of rheumatoid arthritis in adults and an economic evaluation of their cost-effectiveness.Health Technol Assess. 2006 Nov;10(42):iii-iv, xi-xiii, 1-229. doi: 10.3310/hta10420. Health Technol Assess. 2006. PMID: 17049139
-
HTR2A DNA methylation as a diagnostic biomarker for rheumatoid arthritis: a validation study using targeted sequencing.Epigenomics. 2025 Aug;17(11):721-731. doi: 10.1080/17501911.2025.2523231. Epub 2025 Jun 27. Epigenomics. 2025. PMID: 40576051
-
Newborn mitochondrial DNA copy number is associated with changes to DNA methylation that persist into childhood and are associated with cognitive development.Clin Epigenetics. 2025 Jul 2;17(1):112. doi: 10.1186/s13148-025-01896-y. Clin Epigenetics. 2025. PMID: 40605063 Free PMC article.
-
DNA methylation markers associated with type 2 diabetes, fasting glucose and HbA1c levels: a systematic review and replication in a case-control sample of the Lifelines study.Diabetologia. 2018 Feb;61(2):354-368. doi: 10.1007/s00125-017-4497-7. Epub 2017 Nov 21. Diabetologia. 2018. PMID: 29164275 Free PMC article.
References
-
- Pedersen M, Jacobsen S, Garred P, Madsen HO, Klarlund M, Svejgaard A, et al. Strong combined gene-environment effects in anti-cyclic citrullinated peptide-positive rheumatoid arthritis: A nationwide case-control study in Denmark. Arthritis Rheumatol. (2007) 56:1446–53. doi: 10.1002/art.22597, PMID: - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

