Inverted chimeric RNAi molecules synergistically cotarget MYC and KRAS in KRAS-driven cancers
- PMID: 40762837
- PMCID: PMC12483557
- DOI: 10.1172/JCI187204
Inverted chimeric RNAi molecules synergistically cotarget MYC and KRAS in KRAS-driven cancers
Abstract
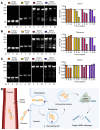
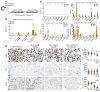
Mutant KRAS has been implicated in driving a quarter of all cancer types. Although inhibition of the KRASG12C mutant protein has shown clinical promise, there is still a need for therapies that overcome resistance and target non-KRASG12C mutations. KRAS activates downstream MYC, which is also a difficult-to-drug oncoprotein. We have developed an "inverted" RNAi molecule with the passenger strand of a MYC-targeting siRNA fused to the guide strand of a KRAS-targeting siRNA. The chimeric molecule simultaneously inhibits KRAS and MYC, showing marked improvements in efficacy beyond the individual siRNA components. This effect is mediated by 5'-dT overhangs following endosomal metabolism. The synergistic RNAi activity led to a more than 10- to 40-fold improvement in inhibition of cancer viability in vitro. When conjugated to an EGFR-targeting ligand, the chimeric siRNA was delivered to and internalized by tumor cells. As compared with individual targeting siRNAs, the chimeric design resulted in considerably improved metabolic stability in tumors, enhanced silencing of both oncogenes, and reduced tumor progression in multiple cancer models. This inverted chimeric design establishes proof of concept for ligand-directed, dual silencing of KRAS and MYC in cancer and constitutes an innovative molecular strategy for cotargeting any two genes of interest, which has broad implications.
Keywords: Drug therapy; Lung cancer; Oncogenes; Oncology; Therapeutics.
Conflict of interest statement
Figures







References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

