METTL1 promotes cadmium-induced stress granules formation via enhancing translation of G3BP1 and expression of m7G- 3' tiRNA MetCAT
- PMID: 40762925
- PMCID: PMC12325392
- DOI: 10.1007/s10565-025-10072-0
METTL1 promotes cadmium-induced stress granules formation via enhancing translation of G3BP1 and expression of m7G- 3' tiRNA MetCAT
Abstract
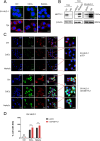
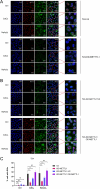
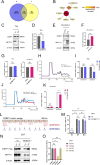
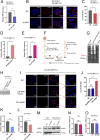
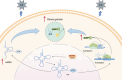
Methyltransferase 1 (METTL1) is currently regarded as a key tRNA m7G writer. Recent studies indicate its potential role in carcinogenesis via increased m7G modification to stabilize tRNA and upregulate tRNA expression. Cadmium-induced stress triggers the assembly of stress granules (SGs) and production of tRNA-derived stress-induced RNAs (tiRNAs). However, whether METTL1 is involved in the formation of cadmium-induced SGs and its mechanism are still unclear. Here, we demonstrated that METTL1 promotes cadmium-induced SGs formation. Mechanistically, METTL1 not only upregulates SG's core protein Ras-GTPase-activating protein SH3 domain-binding protein 1 (G3BP1) translation through tRNAs m7G modification, but also enhances expression of one m7G-modified tiRNA, m7G-3' tiRNA-MetCAT (mtiRM), which affects SGs assembly. Together, the findings concluded that the promotional effect of METTL1 on cadmium-induced SGs formation jointly through G3BP1 translation and mtiRM expression, thus providing insights into an intimate link between SGs and tumorigenesis.
Keywords: Cadmium; G3BP1; METTL1; Stress granules; TsRNAs.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: The authors declare no competing interests.
Figures

References
-
- Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–6. 10.1038/nrm2694. - PubMed
-
- Andrews SR. FastQC: a quality control tool for high throughput sequence data.; 2010.
MeSH terms
Substances
Grants and funding
- No. 82373427, No. 82072939/National Natural Science Foundation of China
- No. 82272995, No. 8247264, No. 82073047/National Natural Science Foundation of China
- No. 202206010110/Guangzhou Science and Technology Program key projects
- No. 2022A1515010425/Guangdong Basic and Applied Basic Research Foundation
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

