BRD4 inhibition leads to MDSC apoptosis and enhances checkpoint blockade therapy
- PMID: 40762981
- PMCID: PMC12483567
- DOI: 10.1172/JCI181975
BRD4 inhibition leads to MDSC apoptosis and enhances checkpoint blockade therapy
Abstract
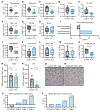
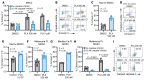
BRD4 is an epigenetic reader protein that regulates oncogenes such as myc in cancer. However, its additional role in shaping immune responses via regulation of inflammatory and myeloid cell responses is not yet fully understood. This work further characterized the multifaceted role of BRD4 in antitumor immunity. Nanostring gene expression analysis of EMT6 tumors treated with a BRD4 inhibitor identified a reduction in myeloid gene expression signatures. Additionally, BRD4 inhibition significantly reduced myeloid-derived suppressor cells (MDSCs) in the spleens and tumors of mice in multiple tumor models and also decreased the release of tumor-derived MDSC growth and chemotactic factors. Pharmacologic inhibition of BRD4 in MDSCs induced apoptosis and modulated expression of apoptosis regulatory proteins. A BRD4 myeloid-specific knockout model suggested that the dominant mechanism of MDSC reduction after BRD4 inhibition was primarily through a direct effect on MDSCs. BRD4 inhibition enhanced anti-PD-L1 therapy in the EMT6, 4T1, and Lewis lung carcinoma tumor models, and the efficacy of the combination treatment was dependent on CD8+ T cells and on BRD4 expression in the myeloid compartment. These results identify BRD4 as a regulator of MDSC survival and provide evidence to further investigate BRD4 inhibitors in combination with immune-based therapies.
Keywords: Apoptosis; Cancer; Cancer immunotherapy; Immunology; Oncology.
Figures





References
-
- Stathis A, Bertoni F. BET proteins as targets for anticancer treatment. Cancer Discov. 2018;8(1):24–36. doi: 10.1158/2159-8290.CD-17-0605. - DOI - PubMed
-
- Tasdemir N, et al. BRD4 connects enhancer remodeling to senescence immune surveillance. Cancer Discov. 2016;6(6):612–629. doi: 10.1158/2159-8290.CD-16-0217. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

