Development and Validation of DIANA (Diabetes Novel Subgroup Assessment tool): A web-based precision medicine tool to determine type 2 diabetes endotype membership and predict individuals at risk of microvascular disease
- PMID: 40763148
- PMCID: PMC12324136
- DOI: 10.1371/journal.pdig.0000702
Development and Validation of DIANA (Diabetes Novel Subgroup Assessment tool): A web-based precision medicine tool to determine type 2 diabetes endotype membership and predict individuals at risk of microvascular disease
Abstract
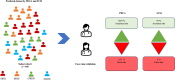
Background: Previous research has identified four distinct endotypes of type 2 diabetes in Asian Indians, which include Severe Insulin Deficient Diabetes (SIDD), Combined Insulin Resistant and Deficient Diabetes (CIRDD), Insulin Resistance and Obese Diabetes (IROD), and Mild Age-related Diabetes (MARD). DIANA (Diabetes Novel Subgroup Assessment) is an online precision medicine tool that can predict endotype membership of type 2 diabetes and individual risk for retinopathy and nephropathy.
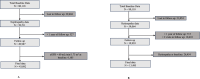
Methodology: The DIANA tool determines subgroup membership using a machine learning model (support vector machine) on T2D subgroups in the Asian Indian population. We used a support vector machine (SVM) model to classify type 2 diabetes patient endotypes, and the model is trained based on k-fold cross-validation. Its performance was compared with an algorithm determined based on conditional pre-determined cut-offs and weights for each clinical feature [age at diagnosis, BMI, waist, HbA1c, Serum Triglycerides, HDL-Cholesterol, (C-peptide fasting, C-peptide stimulated) - optional. This study employed local interpretable model-agnostic explanations (LIME) and SHapley Additive exPlanations (SHAP) to demystify the endotype prediction model. A random forest model was built to assess an individual's risk for nephropathy and retinopathy based on individual risk algorithms.
Findings: The SVM model has relatively high accuracy, specificity, sensitivity, and precision values compared to conditional pre-determined cut-offs 98% vs 63.6%, 99.8% vs 88%, 98.5% vs 65.1%, and 98.7% vs 63.4%. Clinician face value validation of the prediction by the SVM model reported an accuracy, specificity, sensitivity and precision compared to conditional pre-determined cut-offs 97% vs 85%, 95.3% vs 63%, 95.8% vs 73%, and 98.9% vs 66.9%. Additionally, our study demonstrated the impact of features on ML models through LIME and SHAP analyses. The accuracy of the random forest risk prediction model for nephropathy and retinopathy was 89.6% (p < 0.05) and 78.4% (p < 0.05), respectively.
Conclusion: We conclude that, DIANA is an accurate, clinically explainable AI tool that clinicians can use to make informed decisions on risk assessment and provide precision management to individuals with new-onset type 2 diabetes.
Copyright: © 2025 Baskar et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
A Responsible Framework for Assessing, Selecting, and Explaining Machine Learning Models in Cardiovascular Disease Outcomes Among People With Type 2 Diabetes: Methodology and Validation Study.JMIR Med Inform. 2025 Jun 27;13:e66200. doi: 10.2196/66200. JMIR Med Inform. 2025. PMID: 40577645 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Prediction of Insulin Resistance in Nondiabetic Population Using LightGBM and Cohort Validation of Its Clinical Value: Cross-Sectional and Retrospective Cohort Study.JMIR Med Inform. 2025 Jun 13;13:e72238. doi: 10.2196/72238. JMIR Med Inform. 2025. PMID: 40512995 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Quality improvement strategies for diabetes care: Effects on outcomes for adults living with diabetes.Cochrane Database Syst Rev. 2023 May 31;5(5):CD014513. doi: 10.1002/14651858.CD014513. Cochrane Database Syst Rev. 2023. PMID: 37254718 Free PMC article.
References
-
- Atlas G. Diabetes. International diabetes federation. IDF Diabetes Atlas; 2021.
-
- Mohan V, Deepa M, Anjana RM, Lanthorn H, Deepa R. Incidence of diabetes and pre-diabetes in a selected urban South Indian population (CUPS-19). J Assoc Physic India. 2008;56:152–7. - PubMed
-
- Mohan V, Ramachandran A, Snehalatha C, Mohan R, Bharani G, Viswanathan M. High prevalence of maturity-onset diabetes of the young (MODY) among Indians. Diabetes Care. 1985;8(4):371–4. - PubMed
-
- Anjana RM, Baskar V, Nair AT, Jebarani S, Siddiqui MK, Pradeepa R, et al. Novel subgroups of type 2 diabetes and their association with microvascular outcomes in an Asian Indian population: a data-driven cluster analysis: the INSPIRED study. BMJ Open Diabetes Research & Care. 2020;8:e001506. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
