Comparative assessment of line probe assays and targeted next-generation sequencing in drug-resistant tuberculosis diagnosis
- PMID: 40763426
- PMCID: PMC12344966
- DOI: 10.1016/j.ebiom.2025.105875
Comparative assessment of line probe assays and targeted next-generation sequencing in drug-resistant tuberculosis diagnosis
Abstract
Background: Rapid and accurate detection of drug-resistant tuberculosis (DR-TB) is crucial for ensuring effective treatment, halting transmission and preventing the amplification of resistance. Comparative evaluations of molecular diagnostic assays in high-burden settings are essential for informing clinical decision-making for DR-TB treatment.
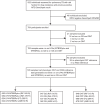
Methods: The Seq&Treat clinical study previously evaluated the performance of two targeted next-generation sequencing (tNGS) workflows, GenoScreen Deeplex Myc-TB and Oxford Nanopore Technologies Tuberculosis Drug Resistance Test, on direct sediment samples from persons at risk for DR-TB. Hain Line Probe Assay (LPAs-MTBDRplus and MTBDRsl) were run as a comparator test using an aliquot of the same sediment samples. Diagnostic performance of the LPAs and previously established tNGS performance were compared, including sensitivity and specificity, for rifampicin, isoniazid, fluoroquinolones (moxifloxacin, levofloxacin), and amikacin, using a composite reference standard of phenotypic drug susceptibility testing and whole-genome sequencing.
Findings: Among 720 clinical samples tested, MTBDRplus LPA sensitivity for rifampicin and isoniazid was 92.3% (95% CI 88.9-94.8) and 91.9% (88.4-94.4), each significantly lower than ≥95% achieved by both tNGS workflows (p < 0.01). For fluoroquinolones (moxifloxacin and levofloxacin), the MTBDRsl LPA and ONT had similar sensitivities (94.3% and 92.7%, and 94.8% and 93.9%, respectively), while GenoScreen outperformed both (97.3% and 96.6%). GenoScreen also demonstrated the highest sensitivity for amikacin resistance (94.6%) compared to LPAs (88.7%) and ONT (88.3%). Complete assay failure rates were low for LPAs (4.9%) and ONT (5.0%) and moderately higher for GenoScreen (8.6%), with differences in single-target failures across all assays.
Interpretation: LPAs demonstrated lower sensitivity and more limited drug resistance detection compared to tNGS workflows, underscoring the advantages of tNGS for improving DR-TB diagnostic algorithms. These findings provide critical evidence to guide updates in DR-TB diagnostic programs.
Funding: Support for the Seq&Treat project was provided through funding from Unitaid (2019-32-FIND MDR).
Keywords: Drug resistance; Line probe assay; Sequencing; Targeted NGS; Tuberculosis; tNGS.
Copyright © 2025 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests TCR, MS, REC received salary support from FIND through a service contact to UC San Diego. TCR and REC Received grant funding from NIH to develop and evaluate a tNGS solution for drug resistant TB (R01AI176401). TCR and REC are co-inventors on a patent associated with the processing of TB sequencing data (European Patent Application No. 14840432.0 & USSN 14/912,918). Both TCR and REC have transferred all rights and present and future interest in and rights to royalties from this patent to UC San Diego and Translational Genomics Research Institute respectively. TCR is a co-founder, board member and unpaid shareholder of Verus Diagnostics Inc, a company that was founded with the intent of developing diagnostic assays. Verus Diagnostics is not pursuing any drug resistant TB diagnostics nor any diagnostics related to the technology or approaches discussed or mentioned in this manuscript. Verus Diagnostics was not involved in any way with data collection, analysis or publication of the results of this manuscript. TCR has not received any financial support from Verus Diagnostics. CR has received honoraria payments from Becton Dickinson and she is on the scientific advisory board for Cepheid and bioMérieux. All other authors declare no competing interests.
Figures

References
-
- World Health Organization . 2023. Global tuberculosis report 2023. Geneva.
-
- World Health Organization . World Health Organization; Geneva: 2024. Global tuberculosis report 2024. Contract No.: Licence: CC BY-NC-SA 3.0 IGO.
-
- World Health Organization . 2022. Global tuberculosis report 2022. Geneva.
-
- World Health Organization . Module 3: Diagnosis Rapid Diagnostic for tuberculosis detection. 2021. WHO consolidated guidelines on tuberculosis.
-
- Global Laboratory Initiative . 2022. Line probe assays for detection of drug-resistant tuberculosis: interpretation and reporting manual for laboratory staff and Clinicians. Contract No.: Licence: CC BY-NC-SA 3.0 IGO.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

