Functional Differences Between SIRPα Splice Isoforms
- PMID: 40763927
- PMCID: PMC12324903
- DOI: 10.1111/gtc.70041
Functional Differences Between SIRPα Splice Isoforms
Abstract
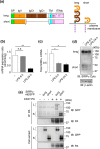
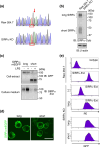
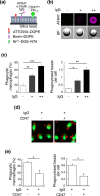
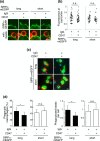
Signal regulatory protein (SIRP) α, an inhibitory receptor belonging to the immunoglobulin (Ig) superfamily is abundantly expressed in phagocytes such as macrophages. CD47, the ligand for SIRPα, is expressed in most healthy cells, and called "don't eat me" signal because it binds to SIRPα on the surface of macrophages and inhibits phagocytosis. SIRPα has multiple splice isoforms, but most functional analyses have been carried out using long SIRPα, the SIRPα isoform with three extracellular Ig domains. In this study, we analyzed the expression and function of short SIRPα, an SIRPα isoform with only one extracellular Ig domain. In resting mouse macrophage Raw 264.7 cells, the short and long SIRPα mRNA expression levels were similar, and the proportion of short SIRPα mRNA decreased substantially after endotoxin stimulation. Short SIRPα bound to CD47 as same as long SIRPα, however, did not suppress the phagocytosis of recombinant CD47-coated beads, unlike long SIRPα. These results suggest that short SIRPα may be a "don't eat me" signal regulator with different expression and function from long SIRPα.
Keywords: CD47; SIRPα; macrophage; phagocytosis; splice isoform; “don't eat me” signal.
© 2025 The Author(s). Genes to Cells published by Molecular Biology Society of Japan and John Wiley & Sons Australia, Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
References
-
- Barclay, A. N. , and Brown M. H.. 2006. “The SIRP Family of Receptors and Immune Regulation.” Nature Reviews. Immunology 6: 457–464. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

