Microhaplotype deep sequencing assays to capture Plasmodium vivax infection lineages
- PMID: 40764298
- PMCID: PMC12325997
- DOI: 10.1038/s41467-025-62357-x
Microhaplotype deep sequencing assays to capture Plasmodium vivax infection lineages
Abstract
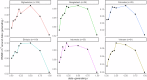
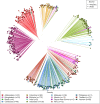
Plasmodium vivax elimination is challenged by dormant liver stages (hypnozoites) that can reactivate months after initial infection resulting in relapses. Relapsing infections confound antimalarial clinical efficacy trials due to the inability to distinguish between recurrences arising from blood-stage treatment failure (recrudescence), reinfection or relapse. Genetic relatedness of paired parasite isolates, measured by identity-by-descent (IBD), can provide important information on whether individuals have had single or multiple mosquito inoculations, thus informing on recurrence origin. We developed a high-throughput amplicon sequencing assay comprising 93 multi-SNP (microhaplotype) markers to determine IBD between P. vivax clinical isolates. The assay was evaluated in 745 global infections, including 128 infection pairs from a randomized controlled trial (RCT) (ClinicalTrials.gov NCT01680406). Simulations demonstrate low error in pairwise IBD estimation at the panel (RMSE < 0.12) and IBD-based networks illustrate strong clustering by geography. IBD analysis in the RCT demonstrates a lower frequency of suspected relapses or recrudescence in patients treated with primaquine compared to those without primaquine; the impact is greater when paired with chloroquine than with artemether-lumefantrine. Our results demonstrate the potential to derive new information on P. vivax treatment and transmission using IBD generated by amplicon sequencing data that can be further improved with time-to-event models.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures





References
-
- WHO. World Malaria Report 2022. World Health Organization; Geneva 2022. (WHO, 2022).
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

