Investigation of ribociclib, abemaciclib and palbociclib resistance in ER+ breast cancer cells reveal potential therapeutic opportunities`
- PMID: 40764324
- PMCID: PMC12325956
- DOI: 10.1038/s41598-025-11052-4
Investigation of ribociclib, abemaciclib and palbociclib resistance in ER+ breast cancer cells reveal potential therapeutic opportunities`
Abstract
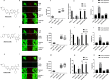
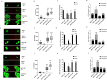
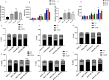
The cyclin-dependent kinase 4/6 inhibitor (CDK4/6i) ribociclib, abemaciclib, and palbociclib have transformed outcomes in patients with ER+ /HER2 - advanced breast cancer (BC). However, most patients eventually progress, and therapeutic options beyond CDK4/6i are an area of ongoing investigation. Here, we generated and evaluated ribociclib, abemaciclib, and palbociclib-resistant BCs. MCF7 and T47D (ER+ /HER2-) cells were chronically treated with increasing doses of ribociclib (R), abemaciclib (A), or palbociclib (P) over 8 months (0-600 nM). CDK4/6i-resistant cell lines (MCF7rR, MCF7rA, MCF7rP, T47DrR, T47DrA, and T47DrP) were isolated and evaluated for their aggressive phenotypes, cross-resistance, transcriptomic changes, and sensitivity to volasertib (PLK1 inhibitor) and barasertib (AukB inhibitor). Immunohistochemical evaluation of CDK4, CDK6, and p53 (n = 1005) and transcriptomic evaluation of AukB and PLK1 were performed in 5031 clinical breast cancers. MCF7rR, MCF7rA, MCF7rP, T47DrR, T47DrA, and T47DrP cells manifested aggressive phenotypes such as increased spheroid formation, invasion, proliferation, and progression through the G1/S phase of the cell cycle despite CDK4/6i treatment, increased resistance to apoptosis, and cross-resistance to other CDK4/6i. Transcriptomic analysis revealed the enrichment of distinct pathways in resistant cells, particularly the upregulation of cell cycle regulatory genes such as PLK1, AukB, CDKN2B and TGFβ. PLK1 or AukB overexpressing resistant cells were sensitive to volasertib (PLK1 inhibitor) and barasertib (AukB inhibitor) therapy, which was associated with G2/M cell cycle arrest and increased apoptosis. We conclude that cell cycle upregulation leading to G2/M progression is a key route for CDK4/6i resistance. AukB or PLK1 inhibitors that block G2/M phase could be a promising strategy.
Keywords: Abemaciclib; Breast cancer; CDK4; CDK6; Mechanism of resistance; Palbociclib; Ribociclib.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Ethics approval: The immunohistochemical study was approved by the Yorkshire & Humber-Leeds East Research Ethics Committee (REC Reference: 19/YH/0293) under IRAS Project ID: 266925. The collected data were anonymized. Patient consent: All patients provided informed written consent.
Figures


Similar articles
-
Real-world effectiveness comparison of first-line palbociclib, ribociclib or abemaciclib plus endocrine therapy in advanced HR-positive/HER2-negative BC patients: results from the multicenter PALMARES-2 study.Ann Oncol. 2025 Jul;36(7):762-774. doi: 10.1016/j.annonc.2025.03.023. Epub 2025 Apr 8. Ann Oncol. 2025. PMID: 40204155
-
Models of Early Resistance to CDK4/6 Inhibitors Unveil Potential Therapeutic Treatment Sequencing.Int J Mol Sci. 2025 Mar 14;26(6):2643. doi: 10.3390/ijms26062643. Int J Mol Sci. 2025. PMID: 40141282 Free PMC article. Clinical Trial.
-
Safety and quality of life of CDK4/6 inhibitors therapy for hormone receptor-positive, human epidermal growth factor receptor 2-negative advanced breast cancer: a multicenter cross-sectional survey in China.BMC Cancer. 2025 May 27;25(1):951. doi: 10.1186/s12885-025-14223-8. BMC Cancer. 2025. PMID: 40426093 Free PMC article.
-
Comparative overall survival of CDK4/6 inhibitors in combination with endocrine therapy in advanced breast cancer.Sci Rep. 2024 Feb 7;14(1):3129. doi: 10.1038/s41598-024-53151-8. Sci Rep. 2024. PMID: 38326452 Free PMC article.
-
Influence of ethnicity on cyclin-dependent kinase inhibitor efficacy and toxicity: A systematic review and meta-analysis.Breast. 2025 Feb;79:103833. doi: 10.1016/j.breast.2024.103833. Epub 2024 Nov 4. Breast. 2025. PMID: 39579620 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

