Genomic insights and metabolic profiling of gut commensal Luoshenia tenuis at strain level
- PMID: 40764512
- PMCID: PMC12325933
- DOI: 10.1038/s41522-025-00793-9
Genomic insights and metabolic profiling of gut commensal Luoshenia tenuis at strain level
Abstract
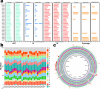
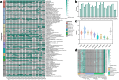
Luoshenia tenuis, a newly identified gut commensal microbe from the family Christensenellaceae, has shown therapeutic effects on weight control and metabolic disorders in model mice. Bacterial strains are essential for investigations on the host-microbe interaction and further development of medical applications. In this study, we collected 27 strains of L. tenuis from the Christensenellaceae Gut Microbial Biobank (ChrisGMB) and sequenced their complete genomes. Our analysis revealed considerable genetic diversity and genomic plasticity. Metabolic prediction indicated that L. tenuis had a preference for metabolizing plant-derived carbohydrates and the ability to synthesize various amino acids and cofactors. In silico analysis, along with in vitro experiments, validated that L. tenuis strains possessed strong acid tolerance and limited antibiotic resistance, suitable traits for oral probiotic development. Further volatile metabolomics and bile acid transformation profiling revealed that L. tenuis was capable of producing metabolites with previously-identified beneficial effects, along with extensive bile acid modification, potentially contributing to its positive impact on host metabolism. This study provides essential insight into strain-level functional and genomic features, laying a foundation for future research towards the development of L. tenuis-based therapies for metabolic disease.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests. Consent for publication: All authors read and agree to the publication of this manuscript.
Figures



References
-
- Fan, Y. & Pedersen, O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol.19, 55–71 (2021). - PubMed
MeSH terms
Substances
Grants and funding
- No.2022YFA1304103/National Key Research and Development Program of China
- No. 32422002/National Natural Science Foundation of China for Excellent Young Scholars
- No. ZR2023YQ020/Shandong Province Natural Science Foundation for Excellent Young Scholars
- No. SKLMTFCP-2023-01/SKLMT Frontiers and Challenges Project
LinkOut - more resources
Full Text Sources

