Ephrin-B2 deletion in GABAergic neurons induces cognitive deficits associated with single-nucleus transcriptomic differences in the prefrontal cortex
- PMID: 40764556
- PMCID: PMC12326787
- DOI: 10.1186/s12915-025-02333-5
Ephrin-B2 deletion in GABAergic neurons induces cognitive deficits associated with single-nucleus transcriptomic differences in the prefrontal cortex
Abstract
Background: Ephrin-B2 (EB2) signaling plays a crucial role in regulating memory and synaptic plasticity. Comprehensive identification of cell-type-specific transcriptomic changes in EB2 knockout mice is expected to shed light on potential mechanisms associated with EB2 signaling in cognitive functions.
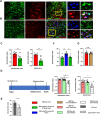
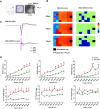
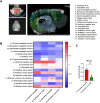
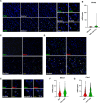
Results: Our study captures changes in cell populations in response to EB2 manipulation and reveals previously uncharacterized cell types (CPA6 + inhibitory neurons) in the mPFC. We validated the differential transcriptomic activity of Pbx1 and Meis1 in CPA6 + neurons using fluorescence in situ hybridization (ISH) in EB2-vGATCre mice. The aberrant presence of CPA6 + neurons in the mPFC may correlate with cognitive impairments induced by EB2 deletion in vGAT + neurons. Analyzing differentially expressed genes (DEGs) in individual cell clusters, we identified alterations related to synapse organization and development, cognition, amyloid-beta formation, and locomotor behavior. Additionally, our DEGs overlapped with human genome-wide association study (GWAS) candidate genes related to cognition and anxiety, underscoring the relevance of our mouse model to human disease.
Conclusions: We present a comprehensive atlas of cell-type-specific gene expression changes in this synaptic deficiency model and identify novel cell-type-specific targets implicated in cognitive deficits. Our investigation provides a detailed map of the cell types, genes, and pathways altered in this inhibitory synaptic deficiency model.
Keywords: Cognition; Ephrin-B2; GABAergic neurons; Single-nucleus RNA sequencing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All experiments involving mice were carried out in accordance with National Institute of Health Guide for the Care and Use of Laboratory Animals and approved by Animal Care and Use Committee in Shanghai Jiao Tong University (IACUC No. A2019102). Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures



Similar articles
-
Cell-type brain-region specific changes in prefrontal cortex of a mouse model of alcohol dependence.Neurobiol Dis. 2024 Jan;190:106361. doi: 10.1016/j.nbd.2023.106361. Epub 2023 Nov 20. Neurobiol Dis. 2024. PMID: 37992784 Free PMC article.
-
Peripuberty Is a Sensitive Period for Prefrontal Parvalbumin Interneuron Activity to Impact Adult Cognitive Flexibility.Dev Neurosci. 2025;47(2):127-138. doi: 10.1159/000539584. Epub 2024 Jun 3. Dev Neurosci. 2025. PMID: 38830346 Free PMC article.
-
Arhgef7 as a key target for enriched environment rescuing spatial cognitive deficits and anxiety-like behaviors in a mouse model of Alzheimer's disease following early social isolation.Alzheimers Res Ther. 2025 Jul 1;17(1):143. doi: 10.1186/s13195-025-01797-5. Alzheimers Res Ther. 2025. PMID: 40598577 Free PMC article.
-
Histone deacetylase 4 inhibition ameliorates the social deficits induced by Ephrin-B2 mutation.Prog Neuropsychopharmacol Biol Psychiatry. 2023 Jan 10;120:110622. doi: 10.1016/j.pnpbp.2022.110622. Epub 2022 Aug 25. Prog Neuropsychopharmacol Biol Psychiatry. 2023. PMID: 36029930
-
ephrin-B2 promotes nociceptive plasticity and hyperalgesic priming through EphB2-MNK-eIF4E signaling in both mice and humans.Pharmacol Res. 2024 Aug;206:107284. doi: 10.1016/j.phrs.2024.107284. Epub 2024 Jun 24. Pharmacol Res. 2024. PMID: 38925462 Free PMC article.
References
-
- Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5(10):793–807. - PubMed
-
- Favuzzi E, Rico B. Molecular diversity underlying cortical excitatory and inhibitory synapse development. Curr Opin Neurobiol. 2018;53:8–15. - PubMed
-
- Holland SJ, Gale NW, Mbamalu G, Yancopoulos GD, Henkemeyer M, Pawson T. Bidirectional signalling through the EPH-family receptor Nuk and its transmembrane ligands. Nature. 1996;383(6602):722–5. - PubMed
MeSH terms
Substances
Grants and funding
- 23ZR1454600/the Natural Science Foundation of Shanghai, China
- SZB2023204/Open Project Funded by the Shanghai Institute of Traditional Chinese Medicine for Mental Health
- xhkp-HM-2024017/Science Popularization and Innovation Project in Xuhui District, Shanghai
- CHDI-2023-B-19/Project for China Hospital Development Institute, Shanghai Jiao Tong University
- 82071500/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources

