Explore the potential mechanism of Huachansu injection against osteosarcoma via metabolomics, network pharmacology and bioinformatics
- PMID: 40764575
- PMCID: PMC12323198
- DOI: 10.1186/s13020-025-01179-x
Explore the potential mechanism of Huachansu injection against osteosarcoma via metabolomics, network pharmacology and bioinformatics
Abstract
Aim: Huachansu injection (HCSI) shows effective medicinal functions against osteosarcoma. This study aimed to reveal the underlying mechanisms of HCSI against osteosarcoma by integrating metabolomics, network pharmacology and bioinformatics.
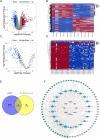
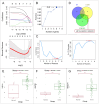
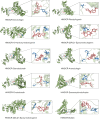
Methods: Metabolomics was used to identify different metabolites and pathways. Network pharmacology was utilized to predict the potential targets of HCSI against osteosarcoma. Differentially expressed lncRNAs and miRNAs were screened and the corresponding lncRNAs-miRNAs-mRNAs network were constructed through the GEO database and miRcode database. Machine learning and immune infiltration analysis were performed on the key target obtained from the intersection of network pharmacology and bioinformatics. The binding affinity between active compounds of HCSI and potential targets was evaluated by molecular docking. The underlying mechanisms were further validated by RT-qPCR and immunoblotting.
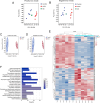
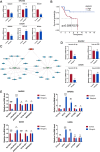
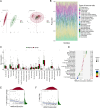
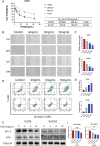
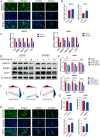
Results: Lipid metabolism pathways were obtained by non-target metabolomics enrichment. A total of 44 HCSI targets associated with osteosarcoma were collected by network pharmacology. Intersection of the mRNAs obtained from ceRNA network with the above 44 targets yielded eight common targets. The main target HMGCR were obtained by machine learning and RT-qPCR. The BCYRN1-miR-27a-3p-HMGCR axis was subsequently screened as the primary ceRNA regulatory network in HSCI against osteosarcoma. Molecular docking also showed an excellent affinity between the active compounds of HCSI and HMGCR. In vitro experiments demonstrated that HCSI down-regulated HMGCR, thereby reduced intracellular cholesterol levels, and ultimately promoting osteosarcoma cell apoptosis.
Conclusion: HCSI could inhibit osteosarcoma progression by regulating lipid metabolism through BCYRN1-miR-27a-3p-HMGCR axis, indicating that HCSI may provide insights for developing herbal medicine injection-based therapies for osteosarcoma.
Keywords: BCYRN1-miR-27a-3p-HMGCR axis; Huachansu injection; Lipid metabolism; Metabolomics; Network pharmacology; Osteosarcoma.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: The manuscript is approved by all authors for publication. Competing interests: The authors have declared no Competing interests.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources

