Decoding Proteus mirabilis biofilms: expression of selected virulence genes and association with antibiotic resistance
- PMID: 40764909
- PMCID: PMC12323196
- DOI: 10.1186/s12866-025-04212-z
Decoding Proteus mirabilis biofilms: expression of selected virulence genes and association with antibiotic resistance
Abstract
Background: Proteus mirabilis is a uropathogens with a strong ability to form resilient crystalline biofilms, particularly on urinary catheters, contributing to its persistence and antibiotic resistance. As biofilm-driven virulence is key in complicated urinary tract infections, understanding its virulence genes and resistance mechanisms is crucial for improving treatment strategies. We investigated the presence, and expression of key virulence genes (ureC, mrpA, speA, and rsbA) in biofilm-forming P. mirabilis strains sourced from both urine (n = 26) and non-urine specimens, such as pus, wounds, and blood isolates (n = 26) and analyzed their association with antimicrobial resistance profiles. The presence and expression of P.mirabilis's virulence genes were detected using conventional PCR and Quantitative real-time PCR assays (qPCR), respectively. Antibiotic susceptibility test (AST) was conducted using the Kirby-Bauer method, adhering to Clinical and Laboratory Standards Institute (CLSI) guidelines. Statistical analysis was performed using the R language.
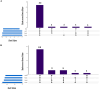
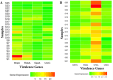
Results: Virulence genes (ureC, mrpA, speA, and rsbA) exhibited high prevalence (> 92% in urine, > 84% in non-urine isolates) with no significant differences (Cochran's Q test; p = 0.801). Multidrug resistance (MDR) and extensively drug-resistant (XDR) were detected in 100% and 57.69% of urine isolates, and 96.15% and 65.38% of non-urine isolates, respectively. Gene combinations are strongly linked to higher resistance rates. mrpA exhibited the highest expression in urine-derived strains, followed by rsbA, and ureC, with speA having the lowest. Post-hoc analysis revealed a significant variation in the rsbA expression compared to speA (p = 0.029) and ureC (p = 0.007). speA showed the highest expression in non-urine isolates, followed by rsbA, ureC, and mrpA, with significant differences among all gene pairs (Conover's all-pairs test; p > 0.05) except rsbA vs. speA. XDR status showed no significant effect or interaction on gene expression in urine and non-urine isolates (p = 0.290; p > 0.05).
Conclusion: The virulence genes ureC, mrpA, speA, and rsbA are consistently found in P. mirabilis strains from both urine and non-urine specimens; however, their expression varies significantly, likely due to host or environmental factors. The presence of high multidrug-resistant (MDR) and extensively drug-resistant (XDR) P. mirabilis strains, potentially driven by the combination of these virulence genes, suggests an increase in virulence.
Keywords: Antibiotic resistance; Biofilm forming Proteus mirabilis; Expression; Genes; Urinary tract infections; Virulence.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests. Clinical trial number: Not applicable.
Figures

Similar articles
-
Characterisation of Proteus mirabilis isolates from the poultry production chain in Zhejiang Province, China: antimicrobial resistance, virulence factors and genotypic profiling.Br Poult Sci. 2025 Aug;66(4):505-514. doi: 10.1080/00071668.2024.2436995. Epub 2025 Jan 24. Br Poult Sci. 2025. PMID: 39853207
-
Biofilm formation ability and swarming motility are associated with some virulence genes in Proteus mirabilis.BMC Microbiol. 2025 Jul 2;25(1):388. doi: 10.1186/s12866-025-04090-5. BMC Microbiol. 2025. PMID: 40604381 Free PMC article.
-
Outbreak Caused by VIM-1- and VIM-4-Positive Proteus mirabilis in a Hospital in Zagreb.Pathogens. 2025 Jul 26;14(8):737. doi: 10.3390/pathogens14080737. Pathogens. 2025. PMID: 40872247 Free PMC article.
-
Point-of-care tests for urinary tract infections to reduce antimicrobial resistance: a systematic review and conceptual economic model.Health Technol Assess. 2024 Nov;28(77):1-109. doi: 10.3310/PTMV8524. Health Technol Assess. 2024. PMID: 39644102 Free PMC article.
-
The diagnostic accuracy of the GenoType(®) MTBDRsl assay for the detection of resistance to second-line anti-tuberculosis drugs.Cochrane Database Syst Rev. 2014 Oct 29;(10):CD010705. doi: 10.1002/14651858.CD010705.pub2. Cochrane Database Syst Rev. 2014. PMID: 25353401 Free PMC article.
References
-
- Stickler D, Morris N, Moreno MC, Sabbuba N. Studies on the formation of crystalline bacterial biofilms on urethral catheters. Eur J Clin Microbiol Infect Dis. 1998;17:649–52. - PubMed
-
- Yuan F, Huang Z, Yang T, Wang G, Li P, Yang B, et al. Pathogenesis of Proteus mirabilis in catheter-associated urinary tract infections. Urol Int. 2021;105(5–6):354–61. - PubMed
-
- Hussein EI, Al-Batayneh K, Masadeh MM, Dahadhah FW, Al Zoubi MS, Aljabali AA, et al. Assessment of pathogenic potential, virulent genes profile, and antibiotic susceptibility of Proteus mirabilis from urinary tract infection. Int J Microbiol. 2020;2020(1):1231807. 10.1155/2020/1231807. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

