Drug repurposing for Alzheimer's disease using a graph-of-thoughts based large language model to infer drug-disease relationships in a comprehensive knowledge graph
- PMID: 40764997
- PMCID: PMC12326721
- DOI: 10.1186/s13040-025-00466-5
Drug repurposing for Alzheimer's disease using a graph-of-thoughts based large language model to infer drug-disease relationships in a comprehensive knowledge graph
Abstract
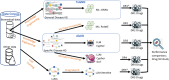
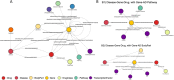
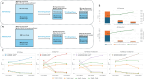
Drug repurposing (DR) offers a promising alternative to the high cost and low success rate of traditional drug development, especially for complex diseases like Alzheimer's disease (AD). This study addressed DR for AD from three key angles: (1) demonstrating how disease-specific knowledge graphs can improve DR performance, (2) evaluating the role of large language models (LLMs) in enhancing the usability and efficiency of these graphs, and (3) assessing whether Graph-of-Thoughts (GoT)-enhanced LLMs, when integrated with AD knowledge graphs, can outperform traditional machine learning and LLM-based approaches. We tested five distinct DR strategies (DR1-DR5) for AD: DR1, a machine learning method using TxGNN; DR2, a machine learning model leveraging the Alzheimer's KnowledgeBase (AlzKB); DR3, an LLM-based chatbot built on AlzKB; DR4, our ESCARGOT framework combining GoT-enhanced LLMs with AlzKB; and DR5, a general reasoning-driven LLM approach. Results showed that AlzKB significantly improved DR outcomes. ESCARGOT further enhanced performance while reducing the need for coding or advanced expertise in knowledge graph analysis. Because the architecture of AlzKB is easily adaptable to other diseases and ESCARGOT can integrate with various knowledge graph platforms, this framework offers a broadly applicable, innovative tool for accelerating drug discovery through repurposing.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: This research used publicly available datasets and did not require additional ethical approval or consent to participate. Consent for publication: All authors have reviewed the final version of the manuscript and have given their full consent for its publication. Competing interests: Dr. Jason H. Moore is the Editor-in-Chief of Biodata Mining and a co-editor of the topical collection “Advances in Data Mining for Biomedical Informatics and Healthcare”.
Figures
Similar articles
-
Leveraging Medical Knowledge Graphs Into Large Language Models for Diagnosis Prediction: Design and Application Study.JMIR AI. 2025 Feb 24;4:e58670. doi: 10.2196/58670. JMIR AI. 2025. PMID: 39993309 Free PMC article.
-
The Alzheimer's Knowledge Base: A Knowledge Graph for Alzheimer Disease Research.J Med Internet Res. 2024 Apr 18;26:e46777. doi: 10.2196/46777. J Med Internet Res. 2024. PMID: 38635981 Free PMC article.
-
Enhancing Pulmonary Disease Prediction Using Large Language Models With Feature Summarization and Hybrid Retrieval-Augmented Generation: Multicenter Methodological Study Based on Radiology Report.J Med Internet Res. 2025 Jun 11;27:e72638. doi: 10.2196/72638. J Med Internet Res. 2025. PMID: 40499132 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
-
- Besta M, Blach N, Kubicek A, Gerstenberger R, Podstawski M, Gianinazzi L, et al. Graph of Thoughts: Solving Elaborate Problems with Large Language Models. Proceedings of the AAAI Conference on Artificial Intelligence. 2024;38(16):17682–17690. Number: 16. 10.1609/aaai.v38i16.29720.
-
- Bonner S, Barrett IP, Ye C, Swiers R, Engkvist O, Hoyt CT, et al. Understanding the performance of knowledge graph embeddings in drug discovery. Artif Intell Life Sci. 2022;2:100036. 10.1016/j.ailsci.2022.100036.
Grants and funding
LinkOut - more resources
Full Text Sources

