Detecting actionable mutations from matched plasma-based versus tissue next-generation sequencing in advanced non-small cell lung cancer: a retrospective single centre analysis on site
- PMID: 40765058
- PMCID: PMC12326616
- DOI: 10.1186/s13046-025-03480-x
Detecting actionable mutations from matched plasma-based versus tissue next-generation sequencing in advanced non-small cell lung cancer: a retrospective single centre analysis on site
Abstract
Background: Liquid biopsies (LB) are used increasingly to detect actionable mutations in patients newly diagnosed with advanced non-small cell lung cancer (aNSCLC), though tissue biopsies (TB) still remain the gold standard. The value of systematically combining LB and TB next-generation sequencing (NGS) for genomic profiling in these patients remains controversial.
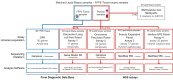
Methods: This single-centre retrospective study included 102 matched TB and LB samples collected from aNSCLC patients at diagnosis. Four circulating free DNA (cfDNA)-based NGS assays (1-4) were compared on site for performance and concordance with TB to detect ESMO Scale for Clinical Actionability of molecular Targets (ESCAT) I/II. Additionally, cfDNA droplet digital PCR methylation (ddPCR-met) testing estimated the tumour fraction to refine the interpretation of wild-type (WT) results.
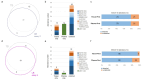
Results: Out of 102 patients, 13% had stage IIIB disease, and 11% presented with brain-only metastases. Adenocarcinoma was the predominant subtype (84%). Ninety LB samples yielded interpretable results across the four assays. Positive percent agreement with TB ranged from 56% (assay 2) to 79% (assay 4), with high concordance, particularly for single-nucleotide variants (SNVs). Hybrid capture-based assays (3 and 4) detected eight and seven gene fusions, respectively, while amplicon-based assays (1 and 2) detected only two each. Assay 3 only identified 12 MET amplifications, five of which were confirmed by fluorescence in situ hybridisation (FISH) but were missed by TB-based NGS. Five out of six negative cfDNA samples with ddPCR-met testing were WT across all assays. The plasma-first approach added incremental value, up to 21% (assay 3). Amplicon-based assays were faster and required less input of DNA for analysis. Patients with stage IIIB or brain-only metastases were significantly more likely to have negative/low levels of cfDNA ddPCR-met.
Conclusions: LB-based NGS demonstrated high concordance with TB in newly diagnosed aNSCLC, particularly for detection of SNV. Hybrid capture assays showed superior performance in identifying gene fusions and MET amplifications. The incremental value of a plasma-first strategy was limited in this real-life study. Thus, LB-based NGS on site should be seen as a complementary tool to TB-based NGS or an alternative when tissue samples are unavailable. Additionally, cfDNA methylation analysis enhances diagnostic accuracy in specific cases.
Keywords: Actionable genomic alterations; CtDNA; Liquid biopsy; Methylation; NGS; Non-small cell lung carcinoma; Precision medicine.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethical approval and consent to participate: All patients signed an informed consent to participate in the study. The study was conducted in accordance with the Declaration of Helsinki, and approved by the ethical committee of Nice Hospital University (NPPS-101188). Competing interests: -Marius IliéHonoraria: AstraZeneca, Merck; Speakers’ Bureau: AstraZeneca; Research Funding: Bristol Myers Squibb, Boehringer Ingelheim; Travel, Accommodations, Expenses: AstraZeneca. -Simon Heeke: Employment: Marker Therapeutics; Honoraria: Qiagen; Consulting or Advisory Role: Boehringer Ingelheim; Speakers’ Bureau: AstraZeneca, Guardant Health; Patents, Royalties, Other Intellectual Property: Patent on the Treatment of Lung Cancer; Travel, Accommodations, Expenses: Roche. -Paul Hofman: Honoraria: Sanofi, Amgen, Roche, BMS, AstraZeneca, AbbVie, Qiagen, Pfizer, Janssen, Pierre Fabre, Thermo Fisher Scientific, Biodena, Diaceutics, Biocartis, Daiichi Sankyo/Lilly, Bayer, Novartis; Consulting or Advisory Role: Sanofi, Amgen, Roche, AbbVie, BMS, AstraZeneca, Pfizer, Janssen, Pierre Fabre, Thermo Fisher Scientific, Qiagen, Diaceutics, Novartis, Biodena, Biocartis, Lilly; Research Funding: Amgen, Thermo Fisher Scientific, Biodena, Roche. No other potential conflicts of interest were reported.
Figures



Similar articles
-
Can a Liquid Biopsy Detect Circulating Tumor DNA With Low-passage Whole-genome Sequencing in Patients With a Sarcoma? A Pilot Evaluation.Clin Orthop Relat Res. 2025 Jan 1;483(1):39-48. doi: 10.1097/CORR.0000000000003161. Epub 2024 Jun 21. Clin Orthop Relat Res. 2025. PMID: 38905450
-
Diagnostic test accuracy and cost-effectiveness of tests for codeletion of chromosomal arms 1p and 19q in people with glioma.Cochrane Database Syst Rev. 2022 Mar 2;3(3):CD013387. doi: 10.1002/14651858.CD013387.pub2. Cochrane Database Syst Rev. 2022. PMID: 35233774 Free PMC article.
-
[Detection of lung cancer driver genes by next-generation sequencing: a comparative analysis of plasma and histological/cytological samples].Zhonghua Bing Li Xue Za Zhi. 2025 Jul 8;54(7):755-761. doi: 10.3760/cma.j.cn112151-20241111-00747. Zhonghua Bing Li Xue Za Zhi. 2025. PMID: 40619265 Chinese.
-
Genomic profiling of NGS-based ctDNA from Chinese non-small cell lung cancer patients.J Cancer Res Clin Oncol. 2023 Sep;149(11):8573-8580. doi: 10.1007/s00432-023-04794-z. Epub 2023 Apr 25. J Cancer Res Clin Oncol. 2023. PMID: 37186065 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 20200108th ed. 2020;70:7–30. - PubMed
-
- Pascual J, Attard G, Bidard FC, Curigliano G, De Mattos-Arruda L, Diehn M et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 20220706th ed. 2022;33:750–68. - PubMed
-
- Mosele F, Remon J, Mateo J, Westphalen CB, Barlesi F, Lolkema MP, et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO precision medicine working group. Ann Oncol. 2020;31:1491–505. - PubMed
-
- Hendriks LE, Kerr KM, Menis J, Mok TS, Nestle U, Passaro A, et al. Oncogene-addicted metastatic non-small-cell lung cancer: ESMO clinical practice guideline for diagnosis, treatment and follow-up. Ann Oncol. 2023;34:339–57. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

