Association of ZNF608 Polymorphisms With House Dust Mite-Induced Allergic Rhinitis
- PMID: 40765419
- PMCID: PMC12326142
- DOI: 10.1002/clt2.70081
Association of ZNF608 Polymorphisms With House Dust Mite-Induced Allergic Rhinitis
Abstract
Background: Genetic factors contribute essentially to the pathophysiology of house dust mite (HDM)-induced allergic rhinitis. Previous studies mainly focused on the biological pathogenesis, but the heritability remains poorly explained.
Methods: A genome-wide gene association analysis (GWGAS) integrating joint-genetic variant effects at the gene level was initially conducted on allergic rhinitis, validated by differential gene expression analysis. A weighted polygenic risk score (wPRS) was used to proxy the cumulative effect of candidate genetic variants in key genes. Gene-set analysis and eQTL analysis were performed to explore the immunologic pathway and genetic regulation of the key gene.
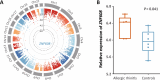
Results: ZNF608 was identified as the key gene involving HDM-induced allergic rhinitis risk (p = 1.23 × 10-6), which was highly expressed in nasal epithelium cells of allergic rhinitis patients (p = 0.041). Furthermore, a wPRS of five significant variants, rs6862252, rs10067299, rs10042766, rs6866116, and rs79679768 in the ZNF608, showed the cumulative effect was associated with the increased HDM-induced allergic rhinitis risk (odds ratio [OR] = 1.40, 95% confidence interval [CI] = 1.18-1.65, p = 1.18 × 10-4), with varied effects under diverse conditions of nasal symptoms. Additionally, both rs6862252 G allele and rs10042766 T allele elevated the expression of ZNF608 involving in state and perturbation of immune cells, such as B cell, T cell, and dendritic cell, contributing to HDM-induced allergic rhinitis.
Conclusion: This study highlights the key gene ZNF608 of HDM-induced allergic rhinitis, which may lay the groundwork for risk assessment and early diagnosis of allergic rhinitis.
Keywords: ZNF608; allergic rhinitis; genetic variants; immune cell; polygenic risk score.
© 2025 The Author(s). Clinical and Translational Allergy published by John Wiley & Sons Ltd on behalf of European Academy of Allergy and Clinical Immunology.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
House dust mite avoidance measures for perennial allergic rhinitis.Cochrane Database Syst Rev. 2001;(4):CD001563. doi: 10.1002/14651858.CD001563. Cochrane Database Syst Rev. 2001. Update in: Cochrane Database Syst Rev. 2007 Jan 24;(1):CD001563. doi: 10.1002/14651858.CD001563.pub2. PMID: 11687117 Updated.
-
House dust mite avoidance measures for perennial allergic rhinitis.Cochrane Database Syst Rev. 2007 Jan 24;(1):CD001563. doi: 10.1002/14651858.CD001563.pub2. Cochrane Database Syst Rev. 2007. Update in: Cochrane Database Syst Rev. 2010 Jul 07;(7):CD001563. doi: 10.1002/14651858.CD001563.pub3. PMID: 17253461 Updated.
-
Saline irrigation for allergic rhinitis.Cochrane Database Syst Rev. 2018 Jun 22;6(6):CD012597. doi: 10.1002/14651858.CD012597.pub2. Cochrane Database Syst Rev. 2018. PMID: 29932206 Free PMC article.
-
Safety and onset time of modified Yupingfeng nasal spray versus mometasone furoate nasal spray on house dust mites-induced moderate to severe allergic rhinitis: A prospective, multicenter, randomized, open-label, parallel-group clinical trial.J Ethnopharmacol. 2025 Mar 26;344:119574. doi: 10.1016/j.jep.2025.119574. Epub 2025 Mar 1. J Ethnopharmacol. 2025. PMID: 40032208 Clinical Trial.
-
House dust mite avoidance measures for perennial allergic rhinitis.Cochrane Database Syst Rev. 2010 Jul 7;2010(7):CD001563. doi: 10.1002/14651858.CD001563.pub3. Cochrane Database Syst Rev. 2010. PMID: 20614426 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources

