Nanog overexpression enhances the therapeutic efficacy of ADMSCs in AMI rats via the upregulation of JAK/STAT3 signaling and cyclin-mitochondrial expression
- PMID: 40765818
- PMCID: PMC12320234
- DOI: 10.7150/ijbs.112824
Nanog overexpression enhances the therapeutic efficacy of ADMSCs in AMI rats via the upregulation of JAK/STAT3 signaling and cyclin-mitochondrial expression
Abstract
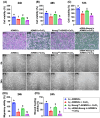
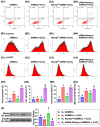
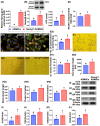
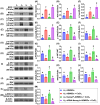
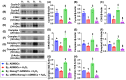
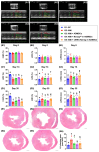
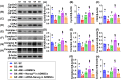
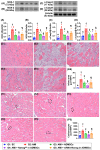
Background: This study investigated whether Nanog-overexpressing adipose-derived mesenchymal stem cells (NanogOE-ADMSCs) are superior to unmodified ADMSCs in improving the left ventricular ejection fraction (LEVF) in acute myocardial infarction (AMI) patients. Methods: We utilized silencing and overexpression of Nanog gene in ADMSCs and performed a wound healing assay/transwell migration assay/MTT cell viability assay/left coronary artery ligation for AMI induction. Additionally, we categorized the cells into three classes [i.e., (ADMSCs and NanogOE-ADMSCs); A1 (ADMSCs)/A2 (ADMSCs + CoCl2)/A3 (NanogOE-ADMSCs + CoCl2)/A4 (siRNA-Nanog-ADMSCs) + CoCl2); B1 (ADMSCs)/B2 (ADMSCs + H2O2)/B3 (NanogOE-ADMSCs + H2O2)/B4 (siRNA-Nanog gene in ADMSCs + H2O2)], and the rats (n=50) were evenly divided into Groups 1 (sham-operated control)/2 (AMI)/3 (AMI+ADMSCs)/4 (AMI+NanogOE-ADMSCs)/5 (AMI+siRNA-Nanog-ADMSCs). The hearts were harvested on Day 35. Results: In vitro experiments revealed significantly higher ATP, relative mitochondrial DNA/Nonog gene expression, mitochondrial cytochrome C+ cell, angiogenesis and exosome-specific marker (Alix/CD81/CD63/CD9) levels in NanogOE-ADMSCs than in ADMSCs. The cell viability, wound healing, and migration were highest in A1, lowest in A4, and significantly greater in A3 than in A2, whereas early/late apoptosis and intracellular and mitochondrial ROS displayed the opposite pattern of cell viability among the groups (all P<0.001). Additionally, the proteins expressions of phosphorylation (p) of the PI3K/Akt/mTOR, p-JAK2/p-STAT3, and Ras/Raf/MEK1/2/ERK1/2 signaling pathways were highest in A3, lowest in A4 and significantly greater in A1 than in A2 (all P<0.001). The levels of cell cycle proteins and mitochondrial electron transport train (ETC) complex I/II/III/IV components exhibited identical patterns as PI3K/Akt/mTOR among the groups B1 to B4 (all P<0.001). On Day 35, the LVEF was highest in Group 1, lowest in Group 2, significantly greater in Group 4 than in Groups 3 and 5, and significantly greater in Group 3 than in Group 5, with the opposite pattern for the LV remodeling index, infarct and fibrosis areas, and LV chamber size (all P < 0.0001). The p-AK/p-STAT3, p-PI3K/p-Akt/p-mTOR, and Ras/Raf/MEK1/2/ERK1/2 protein levels displayed the same pattern as the LVEF among the groups (all P < 0.001). Conclusion: NanogOE-ADMSCs rescued LVEF by upregulating JAK/STAT3-mediated cell proliferation/cell stress pathways and accelerating the cell cycle.
Keywords: Nanog gene overexpression in adipose-derived mesenchymal stem cells; acute myocardial infarction; cell stress signaling.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures


References
-
- Ibanez B, James S, Agewall S, Antunes MJ, Bucciarelli-Ducci C, Bueno H. et al. 2017 ESC Guidelines for the management of acute myocardial infarction in patients presenting with ST-segment elevation: The Task Force for the management of acute myocardial infarction in patients presenting with ST-segment elevation of the European Society of Cardiology (ESC) Eur Heart J. 2018;39:119–77. - PubMed
-
- Antman EM. Time is muscle: translation into practice. J Am Coll Cardiol. 2008;52:1216–21. - PubMed
-
- Redfors B, Mohebi R, Giustino G, Chen S, Selker HP, Thiele H. et al. Time Delay, Infarct Size, and Microvascular Obstruction After Primary Percutaneous Coronary Intervention for ST-Segment-Elevation Myocardial Infarction. Circ Cardiovasc Interv. 2021;14:e009879. - PubMed
-
- Writing Committee M, Gulati M, Levy PD, Mukherjee D, Amsterdam E, Bhatt DL. et al. 2021 AHA/ACC/ASE/CHEST/SAEM/SCCT/SCMR Guideline for the Evaluation and Diagnosis of Chest Pain: A Report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. J Am Coll Cardiol. 2021;78:e187–e285. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

