SIRT4-Mediated Deacetylation of PRDX3 Attenuates Liver Ischemia Reperfusion Injury by Suppressing Ferroptosis
- PMID: 40765819
- PMCID: PMC12320503
- DOI: 10.7150/ijbs.114510
SIRT4-Mediated Deacetylation of PRDX3 Attenuates Liver Ischemia Reperfusion Injury by Suppressing Ferroptosis
Abstract
Liver ischemia-reperfusion injury (LIRI) is an important cause of the clinical prognosis of liver transplantation. Despite Sirtuin 4 (SIRT4) is involved in various post-translational modifications, its role in LIRI is unclear. This research aimed to investigate the influence of SIRT4 on the pathogenesis of LIRI. To this end, SIRT4 knockout (KO) and liver-specific overexpression mice, as well as alpha mouse liver 12 (AML12) cells, were employed. We showed that SIRT4 expression was downregulated in mice with LIRI or AML12 cells exposed to hypoxia-reoxygenation (H/R) injury, as well as in the liver tissue of liver transplant patients. SIRT4 KO exacerbated liver injury and ferroptosis; conversely, liver-specific SIRT4 overexpression in mice produced the opposite results. Furthermore, the ferroptosis inhibitor ferrostatin-1 mitigated the exacerbation of liver injury and ferroptosis caused by SIRT4 KO. Mechanistically, SIRT4 interacted with peroxiredoxins 3 (PRDX3) and deacetylated it at lysine 92, leading to the inhibition of ferroptosis. Furthermore, the protective effect of SIRT4 on LIRI was dependent on PRDX3 deacetylation at lysine 92. Additionally, liver-targeted lipid nanoparticles (LNPs)-sirt4 mRNA alleviated LIRI and ferroptosis in mice. Taken together, our findings highlight the SIRT4-PRDX3 axis as a key regulator and potential therapeutic target for LIRI.
Keywords: PRDX3; SIRT4; deacetylation; ferroptosis; liver ischemia reperfusion injury.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures


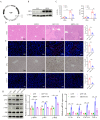

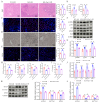




References
-
- Paula Carolina Grande N, João Paulo V, Clarice Fleury F, Karina Dal Sasso M, Maria Cecília Jordani G, Paulo Roberto Barbosa E. et al. Liver ischemia and reperfusion injury. Pathophysiology and new horizons in preconditioning and therapy. Acta Cir Bras. 2018;33:723–35. - PubMed
-
- Kelly M Q, Phillip V B, Luis H T-P. Molecular responses to ischemia and reperfusion in the liver. Arch Toxicol. 2015;89:651–7. - PubMed
-
- Rezà F S, Seyed Kamran Hejazi K. Liver ischemia/reperfusion injury: an overview. J Invest Surg. 2014;27:366–79.
-
- Constantinos N, Konstantinos K, Nikolaos P, Marios-Konstantinos T, Panagis M L, Kassiani T. et al. Global consequences of liver ischemia/reperfusion injury. Oxid Med Cell Longev. 2014;2014:906965.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

