Deciphering the molecular signatures of tropical Areca catechu L. under cold stress: an integrated physiological and transcriptomic analysis
- PMID: 40765861
- PMCID: PMC12321864
- DOI: 10.3389/fpls.2025.1624335
Deciphering the molecular signatures of tropical Areca catechu L. under cold stress: an integrated physiological and transcriptomic analysis
Abstract
Introduction: Areca catechu is a widely cultivated palm species with significant economic and medicinal value. However, A. catechu is a tropical plant that is particularly susceptible to low temperatures.
Methods: This study integrates physiological profiling with transcriptomic sequencing to systematically investigate the cold-response mechanisms of A. catechu.
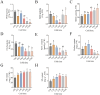
Results: Multivariate variance analysis revealed that peroxidase (POD) activity and chlorophyll content are significant biomarkers strongly correlated with cold tolerance. A comprehensive investigation into the temporal expression of genes in response to 24 hours of cold stress was conducted, using RNA-seq analysis. This analysis yielded a substantial number of differentially expressed genes (DEGs), amounting to 20,870, which were found to be subject to temporal regulation. KEGG pathway enrichment analysis revealed substantial activation in three metabolic pathways: phytohormone signaling, alkaloid biosynthesis (tropane/piperidine/pyridine), and flavonoid biosynthesis. The application of Weighted Gene Co-expression Network Analysis (WGCNA), in conjunction with a dynamic tree-cutting algorithm, resulted in the identification of 25 co-expression modules. Eigenvector centrality analysis identified six hub genes responsive to cold stress: ZMYND15, ABHD17B, ATL8, WNK5, XTH3 and TPS. The findings of this study delineate three key aspects: (1) temporal dynamics of cold-responsive physiological processes, (2) pathway-level characterization of DEG enrichment patterns, and (3) genetic determinants underlying cold stress adaptation.
Discussion: These findings clarify the time series and core physiological indicators of A. catechu during various physiological processes, identify pivotal genes associated with cold stress, and provide a gene-to-phenotype framework for optimizing cold-resilient cultivation protocols and molecular marker-assisted breeding strategies.
Keywords: Areca catechu L.; RNA-seq; WGCNA; cold stress; multivariate analysis.
Copyright © 2025 Li, Zhang, Wen, Ji, Chen, Tian, Yang and He.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures









References
-
- Alam M. U., Fujita M., Nahar K., Rahman A., Anee T. I., Masud A. A. C., et al. (2022). Seed priming upregulates antioxidant defense and glyoxalase systems to conferring simulated drought tolerance in wheat seedlings. Plant Stress 6, 100120. doi: 10.1016/j.stress.2022.100120 - DOI
-
- Alam O., Khan A., Khan W. U., Khan W. A., Ahmad M., Khan L. U. (2024). Improving heat tolerance in betel palm (Areca catechu) by characterisation and expression analyses of heat shock proteins under thermal stress. Crop Pasture Sci. 75. doi: 10.1071/CP24025 - DOI
LinkOut - more resources
Full Text Sources

