Exploring the relationship between co-abundance of gut microbiota and novel metabolic pathways in different subtypes of irritable bowel syndrome: insights from the American Gut Project
- PMID: 40766067
- PMCID: PMC12321851
- DOI: 10.3389/fmed.2025.1615717
Exploring the relationship between co-abundance of gut microbiota and novel metabolic pathways in different subtypes of irritable bowel syndrome: insights from the American Gut Project
Abstract
Background: Irritable bowel syndrome (IBS) is a prevalent functional gastrointestinal disorder with an unclear etiology. Recent studies have underscored the association between alterations in the gut microbiome and the pathogenesis of IBS. However, limited knowledge exists regarding the co-abundance patterns of gut microbiota and metabolic pathways across different IBS subtypes.
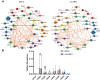
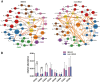
Methods: In this study, we utilized the comprehensive gut microbiome data from the American Gut Project (AGP). Through Spearman correlation analysis, the random forest model, SHAP analysis, and the PICRUSt2 prediction function, we constructed and screened the gut microbiota co-abundance groups and their metabolic characteristics of three cohorts of patients with different subtypes among cohorts of patients with three distinct IBS subtypes: predominant constipation (IBS-C), predominant diarrhea (IBS-D), and unclassified (IBS-U), as well as three non-IBS control groups (non-IBS1, non-IBS2, and non-IBS3, respectively).
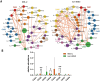
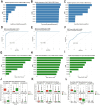
Results: Our study findings indicate that, in comparison to their respective non-IBS groups, there was a significant difference in the prevalence of 37.5% specific co-abundance groups (CAGs) identified across all three IBS subtypes: IBS-C, IBS-D, and IBS-U. In addition, the random forest model shows that there are 2-4 characteristic CAGs for each subtype. We also analyzed the co-abundance networks between each CAG and metabolic pathways. Additionally, we analyzed the co-abundance networks between each CAG and metabolic pathways. No significant species-metabolic pathway co-abundance groups were found in the IBS-C group. In the IBS-D group, 50% of CAGs showed significantly different co-abundance with related metabolic pathways compared to the non-IBS control groups, while in the IBS-U group, this figure was 80%. Through the analysis of differentially expressed metabolic pathways, we revealed significant disturbances in SCFAs and LPS metabolic pathways (particularly a marked increase in acetate) in IBS-D patients, whereas IBS-U patients only exhibited a non-significant downward trend in tryptophan metabolic pathways.
Conclusion: These results indicate that the alterations in the gut microbiota and their associated metabolic pathways differ among IBS subtypes, leading to distinct developments and symptoms. This expands our current understanding of the gut microbiota in different IBS subtypes and provides a theoretical foundation for further research.
Keywords: American Gut Project; co-abundance group; gut microbiota; irritable bowel syndrome subtypes; metabolic pathways.
Copyright © 2025 Han, Mei and Xue.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources

