This is a preprint.
The PLK4 inhibitor RP-1664 demonstrates potent single-agent efficacy in neuroblastoma models through a dual mechanism of sensitivity
- PMID: 40766251
- PMCID: PMC12324582
- DOI: 10.21203/rs.3.rs-7014295/v1
The PLK4 inhibitor RP-1664 demonstrates potent single-agent efficacy in neuroblastoma models through a dual mechanism of sensitivity
Abstract
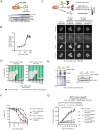
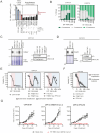
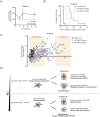
It was recently shown that inhibition of polo-like kinase 4 (PLK4) induces TP53-dependent synthetic lethality in cancers with chromosome 17q-encoded TRIM37 copy number gain due to cooperative regulation of centriole duplication and mitotic spindle nucleation. We show here that chromosome 17q/TRIM37 gain is a pathognomonic feature of high-risk neuroblastoma and renders patient-derived cell lines hypersensitive to the novel PLK4 inhibitor RP-1664. We demonstrate that centriole amplification at low doses of RP-1664 contributes to this sensitivity in a TRIM37- and TP53-independent fashion. CRISPR screens and live cell imaging reveal that upon centriole amplification, neuroblastoma cells succumb to multipolar mitoses due to an inability to cluster or inactivate supernumerary centrosomes. RP-1664 showed robust anti-tumor activity in 14/15 neuroblastoma xenograft models and significantly extended survival in a transgenic murine neuroblastoma model. These data support biomarker-directed clinical development of PLK4 inhibitors for high-risk neuroblastoma and other cancers with somatically acquired TRIM37 overexpression.
Figures




References
-
- Matthay K. K. et al. Neuroblastoma. Nat Rev Dis Primers 2, 16078 (2016). - PubMed
-
- Bown N. et al. Gain of chromosome arm 17q and adverse outcome in patients with neuroblastoma. N Engl J Med 340, 1954–1961 (1999). - PubMed
-
- Gilbert F. et al. Human neuroblastomas and abnormalities of chromosomes 1 and 17. Cancer Res 44, 5444–5449 (1984). - PubMed
-
- Morowitz M. et al. Detection of single-copy chromosome 17q gain in human neuroblastomas using real-time quantitative polymerase chain reaction. Mod Pathol 16, 1248–1256 (2003). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

