Targeting PDCD4 in cancer and atrial fibrillation: mechanistic insights from integrated multi-omics and single-cell analysis
- PMID: 40766329
- PMCID: PMC12321552
- DOI: 10.3389/fonc.2025.1593815
Targeting PDCD4 in cancer and atrial fibrillation: mechanistic insights from integrated multi-omics and single-cell analysis
Abstract
Background: Atrial fibrillation (AF) is a complicated and varied cardiovascular disorder with inadequate understanding of its molecular underpinnings. While Programmed cell death factor 4 (PDCD4) has been associated in several illnesses, its particular significance in AF remains unknown. This work seeks to discover PDCD4-associated critical genes and clarify their regulation processes.
Method: We built a protein-protein interaction (PPI) network to emphasize important biological interactions and used transcriptome analysis to find differentially expressed genes (DEGs). Regulatory mechanisms were explored through miRNA-mRNA and transcription factor (TF) analysis. Single-cell RNA sequencing (SCRNA-SEQ) data were utilized to identify crucial cell types and intercellular communication patterns associated with key genes.
Results: qRT-PCR analysis of peripheral blood mononuclear cells (PBMCs) from AF patients and healthy controls revealed a significant upregulation of PDCD4 in AF patients. Through differential expression analysis and PPI network construction, 11 key genes were identified. In addition, mmu-miR-429-3p regulates Sirt1 while Wt1 shares regulatory roles with PDCD4, Wasl, and Abl2, and that Sirt1 and Atad5 are both regulated by Thap9. Drug prediction analyses revealed sirtinol and trichostatin as promising therapeutic drugs for targeting Atad5 and Sirt1, respectively, with good molecular docking scores (< -5 kcal/mol). SCRNA-SEQ data pinpointed arterial and venous endothelial cells as critical cell types associated with the key genes. Finally, we also found that PDCD4 dysregulation in cancers like ACC may increase AF risk through immune modulation, suggesting that targeting PDCD4 could benefit both AF and ACC patients.
Conclusions: This study demonstrates that PDCD4 modulates AF progression by regulating key genes and pathways involved in inflammation, fibrosis, and metabolic processes. Insights from transcriptome and single-cell analysis give a full knowledge of the molecular processes underlying AF and indicate PDCD4 as a possible therapeutic target.
Keywords: PDCD4; atrial fibrillation; pan-cancer analysis; single-cell RNA sequencing; transcriptome sequencing.
Copyright © 2025 Hailati, Liu, Zhang and Wulasihan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
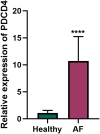



Similar articles
-
Integrated single-cell and transcriptomic analysis of bone marrow-derived metastatic neuroblastoma reveals molecular mechanisms of metabolic reprogramming.Sci Rep. 2025 Aug 5;15(1):28519. doi: 10.1038/s41598-025-13626-8. Sci Rep. 2025. PMID: 40764361 Free PMC article.
-
Comprehensive single-cell chromatin and transcriptomic profiling of peripheral immune cells in nonsegmental vitiligo.Br J Dermatol. 2025 Jun 20;193(1):115-124. doi: 10.1093/bjd/ljaf041. Br J Dermatol. 2025. PMID: 39888372
-
Deciphering Shared Gene Signatures and Immune Infiltration Characteristics Between Gestational Diabetes Mellitus and Preeclampsia by Integrated Bioinformatics Analysis and Machine Learning.Reprod Sci. 2025 Jun;32(6):1886-1904. doi: 10.1007/s43032-025-01847-1. Epub 2025 May 15. Reprod Sci. 2025. PMID: 40374866
-
Intravenous magnesium sulphate and sotalol for prevention of atrial fibrillation after coronary artery bypass surgery: a systematic review and economic evaluation.Health Technol Assess. 2008 Jun;12(28):iii-iv, ix-95. doi: 10.3310/hta12280. Health Technol Assess. 2008. PMID: 18547499
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

