The protease-activated receptors are expressed in glioblastoma and differentially modulate adherent versus stem-like growth of LN-18 GBM cells
- PMID: 40766339
- PMCID: PMC12321897
- DOI: 10.3389/fonc.2025.1582996
The protease-activated receptors are expressed in glioblastoma and differentially modulate adherent versus stem-like growth of LN-18 GBM cells
Abstract
Background: Glioblastoma (GBM) remains the most aggressive and common malignant brain tumor in adults, often accompanied by venous thromboembolism due to hypercoagulability. Protease-activated receptors (PAR1-4) are thought to influence GBM progression, which in this study led to examine their expression in both tissue from GBM patients and in a GBM cell model.
Methods: Using quantitative PCR and immunoblot analyses, we investigated the expression of PAR1-4 in human GBM samples compared to non-malignant brain and evaluated its role in patient survival. In addition, the expression of PAR1-4 in adherent LN-18 GBM cells in comparison to their stem cell-like neurosphere counterparts was analyzed. Finally, the influence of PAR1-4 modulation by specific agonists and antagonists on cell viability was investigated using this GBM cell model.
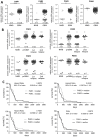
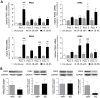
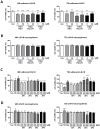
Results: PAR1-4 mRNA levels were significantly higher in GBM than in non-tumoral brain tissue, though this did not affect patient survival. Notably, PAR4 protein levels were lower in GBM, while PAR1, 2, and 3 were unchanged. However, high PAR1 protein levels were linked to poorer patient survival, with a similar trend observed for PAR4, though not statistically significant. Patients with high levels of both PAR1 and PAR3 or PAR4 faced an even greater risk of poor outcomes, but the most severe prognosis was seen in those patients with high PAR3 and PAR4 protein level. In stem-like LN-18 GBM neurospheres, PAR1-4 mRNA was significantly increased, with PAR3 protein elevated and PAR4 reduced. Inhibition of PAR1, PAR2, or PAR4 reduced the viability of adherent GBM cells but not stem-like neurospheres.
Conclusion: These findings suggest that PARs impact GBM patient survival and that tumor stem cells may respond differently to PAR inhibition compared to conventional tumor cells.
Keywords: PARs; glioblastoma; neurospheres; protease activated receptors; stem-like cells.
Copyright © 2025 Bien-Möller, Grober, Albrecht, Paland, Weitmann, Bialke, Marx, Vogelgesang, Tzvetkov, Hoffmann, Schroeder and Rauch.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Pan E, Tsai JS, Mitchell SB. Retrospective study of venous thromboembolic and intracerebral hemorrhagic events in glioblastoma patients. Anticancer Res. (2009) 29:4309–13., PMID: - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

