This is a preprint.
The atypical adhesion GPCR ADGRA1 controls hippocampal inhibitory circuit function
- PMID: 40766348
- PMCID: PMC12324398
- DOI: 10.1101/2025.07.30.667713
The atypical adhesion GPCR ADGRA1 controls hippocampal inhibitory circuit function
Abstract
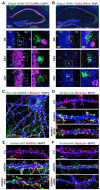
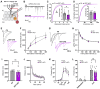
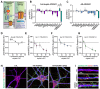
Neural circuits contain a diverse array of inhibitory interneurons that control information processing. The cell surface receptors and signaling pathways that modulate cell type specific inhibitory synaptic function are unclear. Here, we identify the atypical adhesion GPCR ADGRA1 as essential for hippocampal PV and SST inhibitory synaptic function. ADGRA1 is selectively enriched in hippocampal PV and SST interneurons and localizes to a subset of synapses. ADGRA1 deletion in PV and SST interneurons impairs inhibitory synaptic inputs onto Dentate Gyrus granule cells and generates deficits in learning and memory. ADGRA1 engages several downstream G proteins, notably Gα13, a pathway important for the establishment of hippocampal PV interneuron synaptic networks. These results identify an orphan receptor pathway selective for specific inhibitory synapse subtypes and expand our understanding of the signaling mechanisms that establish hippocampal inhibitory circuits.
Conflict of interest statement
Conflict of Interest The authors declare no conflict of interest.
Figures




References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
