This is a preprint.
Typhi Mykrobe: fast and accurate lineage identification and antimicrobial resistance genotyping directly from sequence reads for the typhoid fever agent Salmonella Typhi
- PMID: 40766357
- PMCID: PMC12324182
- DOI: 10.1101/2024.09.30.613582
Typhi Mykrobe: fast and accurate lineage identification and antimicrobial resistance genotyping directly from sequence reads for the typhoid fever agent Salmonella Typhi
Update in
-
Typhi Mykrobe: fast and accurate lineage identification and antimicrobial resistance genotyping directly from sequence reads for the typhoid fever agent Salmonella Typhi.Genome Med. 2025 Oct 24;17(1):130. doi: 10.1186/s13073-025-01551-4. Genome Med. 2025. PMID: 41327441 Free PMC article.
Abstract
Background: Typhoid fever results from systemic infection with Salmonella enterica serovar Typhi (Typhi) and causes 10 million illnesses annually. Disease control relies on prevention (water, sanitation, and hygiene interventions or vaccination) and effective antimicrobial treatment. Antimicrobial resistant (AMR) Typhi lineages have emerged and become established in many parts of the world. Knowledge of local pathogen populations informed by genomic surveillance, including of lineages (defined by the GenoTyphi scheme) and AMR determinants, is increasingly used to inform local treatment guidelines and to inform vaccination strategy. Current tools for genotyping Typhi require multiple read alignment or assembly steps and have not been validated for analysis of data generated with Oxford Nanopore Technologies (ONT) long-read sequencing devices. Here, we introduce Typhi Mykrobe, a command line software tool for rapid genotyping of Typhi lineages, AMR determinants, and plasmid replicons direct from sequencing reads.
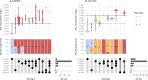
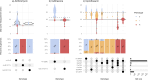
Results: We validated Typhi Mykrobe lineage genotyping by comparison with the current standard read mapping-based approach and demonstrated 99.8% concordance across nearly 13,000 genomes sequenced with Illumina platforms. For the few isolates with discordant calls, we show that Typhi Mykrobe results are better supported by the evidence from raw sequence read data than the results generated using the mapping-based approach. We also demonstrate 99.9% concordance for detection of AMR determinants compared with the current standard assembly-based approach, with similar results for plasmid marker detection. Typhi Mykrobe predicts clinical resistance categorisation (S/I/R) for eight drug classes, and we show strong agreement with phenotypic categorisations generated from reference laboratory minimum inhibitory concentration (MIC) data for n=1,572 Illumina-sequenced isolates (>99% agreement within one doubling dilution). We show strong concordance (>96% for genotype and >98% for AMR and plasmid) between calls made from ONT reads and those made from Illumina reads for isolates sequenced on both platforms (n =93 genomes). Typhi Mykrobe takes less than a minute per sample and is available at https://github.com/typhoidgenomics/genotyphi.
Conclusions: Typhi Mykrobe provides rapid and sensitive genotyping of Typhi genomes direct from Illumina and ONT reads, although lower accuracy was observed for R9 ONT data. It demonstrated accurate assignment of GenoTyphi lineage, detection of AMR determinants and prediction of corresponding AMR phenotypes, and identification of plasmid replicons.
Keywords: Antimicrobial Resistance (AMR); Salmonella Typhi; Typhoid fever; Whole Genome Sequencing (WGS); genotyping; lineage; pathogen sequencing.
Conflict of interest statement
MML was a co-developer of a Trivalent Salmonella (Enteritidis/Typhimurium/Typhi Vi) conjugate vaccine with Bharat Biotech International and the Wellcome Trust. MML has also received payments from Pfizer for consultancy work. MML holds US patents for “Compositions and Methods for Producing Bacterial Conjugate Vaccines”. MML was a member of a NIH DSMB that oversaw US government-funded efficacy trials of COVID-19 vaccines. DSMB was disbanded after several vaccines were given Emergency Use Authorization. MML was a member of the Vaccines and Related Biological Products Advisory Committee of the FDA. IIB was a consultant for the Weapons Threat Reduction Program, Global Affairs Canada. AJP has been involved an Oxford University partnership with AZ for development of COVID-19 vaccines. AJP has received payments for consultancy work from Shionogi. AJP is chair of DHSC’s Joint Committee on Vaccination and Immunisation, is a chair of WHOs Salmonella TAG, and was a member of WHOs SAGE. AJP received support from MRNA – Moderna. KLC has received payments from Pfizer for presentations and travel support from BD. INO has received payments from the Wellcome Trust for consultancy work and receives royalties for books or book chapters published via Springer, Cornell University Press, and Oxford University Press. INO has received travel support from BMGF, ESCMID, and the American ASM and has held leadership or advisory roles for Wellcome SEDRIC, the BMGF surveillance advisory group, the Thomas Bassir Biomedical Foundation, and International Centre for Antimicrobial Resistance Solutions (ICARS) Technical Advisory Forum. ZI has received travel support from ETH Zurich.
Figures


References
-
- Collaborators G 2017 T and P, Stanaway JD, Reiner RC, Blacker BF, Goldberg EM, Khalil IA, et al. The global burden of typhoid and paratyphoid fevers: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect Dis. 2019;19: 369–381. doi: 10.1016/s1473-3099(18)30685-6 - DOI - PMC - PubMed
-
- The WHO AWaRe (Access, Watch, Reserve) antibiotic book. Geneva: World Health Organization; 2022. Licence: CC BY-NC-SA 3.0 IGO.
-
- Collaborators G 2021 F, Vollset SE, Ababneh HS, Abate YH, Abbafati C, Abbasgholizadeh R, et al. Burden of disease scenarios for 204 countries and territories, 2022–2050: a forecasting analysis for the Global Burden of Disease Study 2021. Lancet. 2024;403: 2204–2256. doi: 10.1016/s0140-6736(24)00685-8 - DOI - PMC - PubMed
Publication types
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
